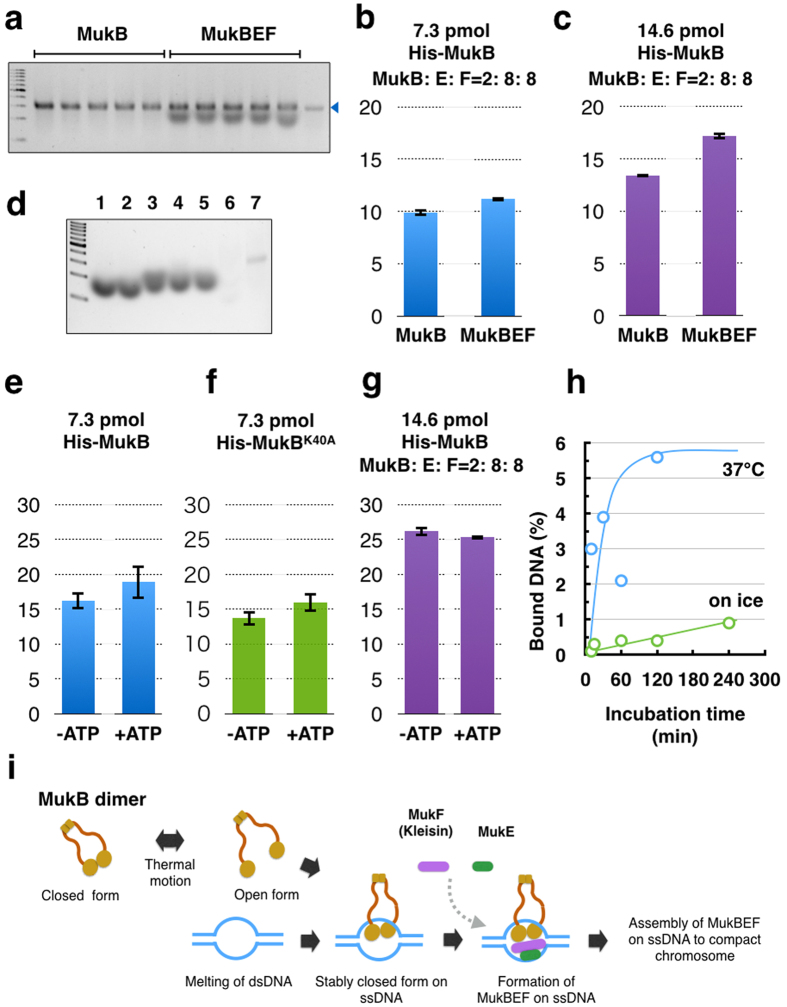
Figure 4. Topological DNA-binding of histidine-tagged MukB in the presence of MukEF and ATP.
(a) Agarose gel electrophoresis of the MU assay. Five reactions with MukB or MukBEF were analyzed and detected by fluorescence of SYBRGreen II. The arrowhead indicates cssDNA of pUC119. (b) Effect of non-SMC subunits, MukE and MukF on retrieved DNA from the MU assay; MukB (7.3 pmol), MukE (29.2 pmol), MukF (29.2 pmol), and pUC119 cssDNA (100 ng). Means and standard deviations were calculated from five independent experiments. (c) Effect of non-SMC subunits, MukE and MukF on retrieved DNA from the MU assay; MukB (14.6 pmol), MukE (58.4 pmol), MukF (58.4 pmol), and pUC119 cssDNA (100 ng). Means and standard deviations were calculated from five independent experiments. (d) Agarose gel electrophoresis of MukF alone. Purified histidine-tagged MukF in HKD buffer was loaded in agarose gel electrophoresis and stained by SYBRGreen II. The results are shown for MukF (30 ng) incubated at 37 °C overnight (lane 1), MukF without incubation (lane 2), MukF incubated with DNase I at 37 °C overnight (lane 3), MukF incubated with RNase A at 37 °C overnight (lane 4), MukF incubated with mung bean nuclease at 37 °C overnight (lane 5), MukF incubated with proteinase K at 37 °C overnight (lane 6), and 10 ng of cssDNA of pUC119 (lane 7). (e) Effect of 2 mM ATP on DNA retrieval from the MU assay. (f) Effect of 2 mM ATP on DNA retrieval of MukBK40A from the MU assay. (g) Effect of 2 mM ATP on DNA retrieval from the MU assay; MukB (14.6 pmol), MukE (58.4 pmol), MukF (58.4 pmol), and pUC119 cssDNA (100 ng). Means and standard deviations were calculated from five independent experiments. (h) Effect of the reaction temperature on DNA retrieval from the MU assay. Reaction mixtures of MukB (7.3 pmol) and pUC119 cssDNA (100 ng) were incubated at 37 °C (blue) and on ice (green). The means and standard deviations (b,d,e) were calculated from five independent experiments. (i) A model of topological binding of MukB in E. coli cells.