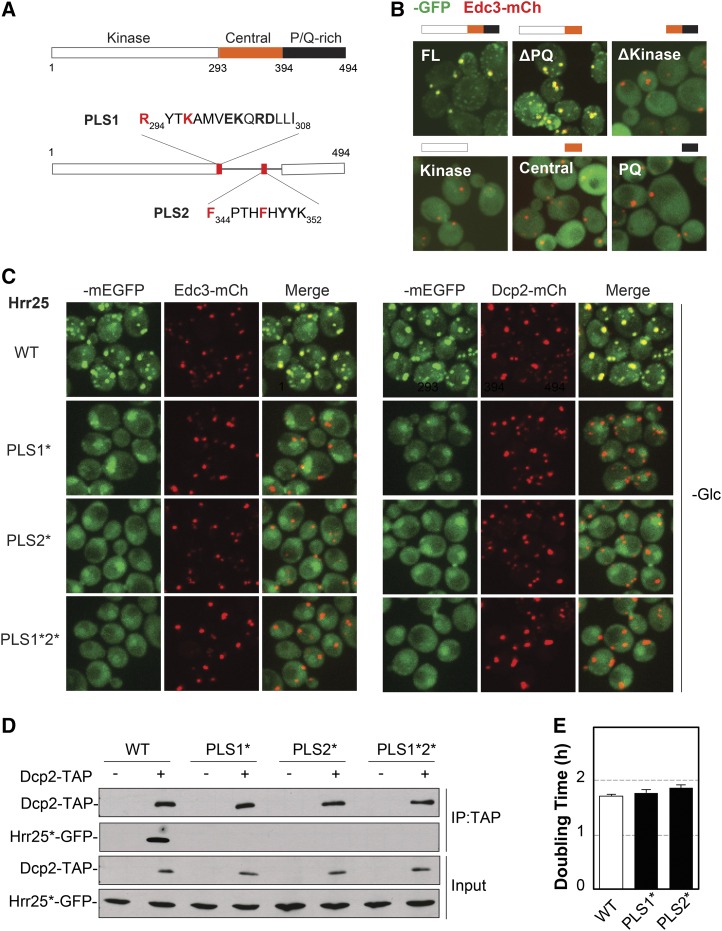
Figure 5.
Two distinct sequence elements in the central domain of Hrr25 were necessary for P-body localization. (A) The domain structure of Hrr25 (top) and the location and sequence of the PLS1 and PLS2 localization signals (bottom). The two residues altered in the PLS1* and PLS* variants are indicated in red. (B) The kinase and central domains of Hrr25 were required for P-body localization. Wild-type cells were transformed with plasmids expressing the indicated GFP-tagged Hrr25 truncation constructs. The localization of these fusion proteins was assessed by fluorescence microscopy after glucose starvation. FL, full-length Hrr25. (C) Alterations in either PLS1 or PLS2 resulted in a loss of Hrr25 localization to P-bodies. Cells expressing the indicated mEGFP-tagged Hrr25 proteins were grown to midlog phase, transferred to a medium lacking glucose, and then visualized by confocal microscopy. (D) Both the PLS1 and PLS2 elements were required for the Hrr25 interaction with Dcp2. TAP-tagged Dcp2 was precipitated from cell extracts and the relative levels of the associated Hrr25 proteins were assessed by Western blotting. (E) Alterations in PLS1 or PLS2 that disrupted P-body localization (PLS1* and PLS2*) did not have a significant effect on the mitotic growth rate. The doubling times shown in the graph are the averages obtained from three independent growth curve determinations.