Abstract
Intersubspecific hybrid sterility is a common form of reproductive isolation in rice (Oryza sativa L.), which significantly hampers the utilization of heterosis between indica and japonica varieties. Here, we elucidated the mechanism of S7, which specially causes Aus-japonica/indica hybrid female sterility, through cytological and genetic analysis, map-based cloning, and transformation experiments. Abnormal positioning of polar nuclei and smaller embryo sac were observed in F1 compared with male and female parents. Female gametes carrying S7cp and S7i were aborted in S7ai/S7cp and S7ai/S7i, respectively, whereas they were normal in both N22 and Dular possessing a neutral allele, S7n. S7 was fine mapped to a 139-kb region in the centromere region on chromosome 7, where the recombination was remarkably suppressed due to aggregation of retrotransposons. Among 16 putative open reading frames (ORFs) localized in the mapping region, ORF3 encoding a tetratricopeptide repeat domain containing protein was highly expressed in the pistil. Transformation experiments demonstrated that ORF3 is the candidate gene: downregulated expression of ORF3 restored spikelet fertility and eliminated absolutely preferential transmission of S7ai in heterozygote S7ai/S7cp; sterility occurred in the transformants Cpslo17-S7ai. Our results may provide implications for overcoming hybrid embryo sac sterility in intersubspecific hybrid rice and utilization of hybrid heterosis for cultivated rice improvement.
Keywords: hybrid sterility, female gamete, tetratricopeptide repeat (TPR), transgenic, rice (Oryza sativa L.)
HYBRIDIZATION between two different species can lead to a distinct phenotype, which can also be fitter than the parental lineage. However, reproductive isolation maintains the integrity of a species over time, reducing or directly impeding gene flow between individuals of different species (Mayr 1942; Grant 1981; Coyne and Orr 2004; Widmer et al. 2009; Baack et al. 2015). The mechanisms of reproductive isolation were classified into two broad categories: prezygotic and postzygotic isolation mechanisms (Mayr 1963; Levin 1978; Sweigart and Willis 2012; Chen et al. 2014). According to the classical Dobzhansky–Muller model, postzygotic isolation results from a deleterious interaction between functionally diverged genes from the hybridizing species (Dobzhansky 1937; Ting et al. 1998; Barbash et al. 2003; Presgraves et al. 2003; Brideau et al. 2006; Bayes and Malik 2009; Ferree and Barbash 2009; Phadnis and Orr 2009; Tang and Presgraves 2009; White et al. 2011). Genes for hybrid sterility, a common pattern of postzygotic isolation, have been reported in several organisms, including fungi, animals, and plants (Brideau et al. 2006; Lee et al. 2008; Bikard et al. 2009; De Vienne et al. 2009).
Major progress has been made in rice and the interspecific and intersubspecific hybrid sterilities are perhaps the best known examples (Chen et al. 2008; Long et al. 2008; Mizuta et al. 2010; Yamagata et al. 2010; Yang et al. 2012). The evolutionary history of rice is complex, but recent work has shed light on the genetics of the transition from wild rice (Oryza rufipogon and O. nivara) to domesticated rice (O. sativa), which comprises two species, Asian rice (O. sativa L.) and African rice (O. glaberrima Steud) (Sweeney and McCouch 2007). The Asian cultivated rice consists of two major types or subspecies, indica and japonica, which are also referred to as “hsien” and “keng,” respectively, since the Han Dynasty (∼2000 years ago) in China (Ting 1957). Recently, a large numbers of loci causing interspecific or intersubspecific hybrid sterility have been identified, including embryo sac abortion (Wan et al. 1993, 1996; Wan and Ikehashi 1995; Zhu et al. 2005; Li et al. 2007; Zhao et al. 2007; Chen et al. 2008, 2012; Yang et al. 2012), pollen sterility (Chen et al. 2006; Jing et al. 2007; Kubo et al. 2008, 2011; Long et al. 2008; Zhang et al. 2011; Zhao et al. 2011), and both in a few cases (Koide et al. 2008, 2012). The S7 locus was first identified causing hybrid sterility between an Aus variety “Ingra” and some javanica varieties by Ikehashi and Araki (1987). Thereafter, S7 was located between Rc (brown pericarp and seed coat) and Est-9 on chromosome 7 (Yanagihara et al. 1992). So far, information is still limited about the molecular mechanism of controlling hybrid embryo sac sterility in rice with S7 locus.
The tetratricopeptide repeat (TPR) motif is a 34 amino acid consensus sequence, commonly found in multiple copies in the same protein. TPR-containing proteins from bacteria to humans have been reported participating in diverse processes such as cell cycle, protein folding, protein kinase inhibition, and hormone regulation (Wang et al. 2004; She et al. 2010; Zeytuni and Zarivach 2012; Li et al. 2015; Masuda et al. 2015). Some TPR-containing proteins, such as FKBP52, TRD-1, and SlTPR1, play important roles in regulation of reproductive development (Tranguch et al. 2005; Yang et al. 2006; Lin et al. 2008; Hughes et al. 2014). In rice, a mutant related to TPR-containing protein (OsAPC6), showed abnormal central cell development during megagametogenesis, leading to failure of endosperm development and reduced seed set (Kumar et al. 2010; Awasthi et al. 2012). However, whether the TPR-containing proteins are involved in hybrid sterility has not been reported.
In this study, we showed that S7 encodes a TPR-containing protein and controls Aus-japonica/indica hybrid female sterility by sporogametophyte allelic interaction. These results are helpful for understanding reproductive isolation and heterosis utilization in rice breeding.
Materials and Methods
Plant materials and growth condition
Ingra with red pericarp belongs to the Aus variety, whereas Cpslo17, possessing a neutral allele at S5 locus, is a javanica variety (Ji et al. 2010) and IR36 is a typical indica variety (Wan and Ikehashi 1996). All plants and populations used for cytological analysis, genetic analysis, fine mapping, and neutral allele identification were planted at the Food Crop Institute, Jiangsu Academy of Agricultural Sciences. Each material was planted with spacing of 16.5 × 16.5 cm. A wide row spacing of 23.5 cm was set between the plots. The plants were managed following normal commercial practices.
Fertility evaluation of pollen, embryo sac, and spikelet
Ten individuals from each parent and F1 hybrid were examined to determine the pollen fertility. Six florets from three panicles of each plant were collected 1–2 days before flowering. One anther per floret per plant was mixed and stained with 1% iodine potassium iodide (I2-KI) solution, and four views were observed by light microscope. The affinity between pollen and stigma was examined by observing the behavior of the pollen grains on the stigma after pollination. Twenty florets were collected at >30 min after flowering to examine the adherence of pollen on stigmata, pollen germination, and elongation of the pollen tube by confocal laser scanning microscopy (Leica TCS SP5). The in vitro pollen germination was tested according to the method of Schreiber and Dresselhaus (2003).
To observe the embryo sac development of the parents and F1 hybrids, spikelets at various development stages were excised and fixed in FAA fluid [containing an 18:1:1 (by volume) mixture of formalin and 70% ethanol and acetic acid]. Before staining, the samples were transferred to 70% ethanol, removing lemma and palea to expose the ovaries. The tissue was then processed through an ethanol series (50, 30, and 15%) and finally transferred into distilled water (20 min each). The whole ovary was incubated in 1 mol/L−1 hydrochloric acid for 15 min, then held in eosin Y water solution for 8 hr, and then in citric acid-disodium hydrogen phosphate buffer (0.1 mol/L−1, pH 5.0) for another 8 hr after washing with distilled water. The ovaries were dyed with 20 μg/ml−1 H33342 (Hoechest stain) for 24 hr at 25° in the dark, washed with distilled water three or four times, and then dehydrated by passing through an ethanol series (15, 30, 50, 70, 85, and 95%) (20 min each) and absolute ethyl alcohol three times (2 hr each), transitted to a mixture of absolute ethyl alcohol and methyl salicylate (1:1) for 1 hr, and cleared in methyl salicylate three times (2 hr each in the previous two steps, and last step >15 hr) (Dai et al. 2006). Fertility of embryo sacs was examined by confocal laser scanning microscopy (Leica TCS SP5).
Spikelet fertility of each plant was determined by counting fertile and sterile spikelets on the upper half of three panicles after maturation as described by Wan et al. (1996).
Transmission ratio distortion and recombination rate
The transmission ratio distortion (TRD) system was measured by the method of Koide et al. (2008). Based on this system, the degree of TRD was calculated in terms of a k value, varying from 0.5 (Mendelian segregation) to 1.0 (complete elimination of its allelic alternative). The km and kf stood for the proportion of progeny that received the allele exhibiting the preferential transmission through male and female gametes, which were estimated from Ns/(Nf + Ns) backcrossing data using heterozygotes as male and female parents, respectively. While Ns represents the number of semisterile plants (heterozygotes Ingra/Cpslo17, S7ai/S7cp or Ingra/IR36, S7ai/S7i), Nf represents fertile plants (homozygotes S7cp/S7cp or S7i/S7i).
Recombination frequency in units of physical distance was defined as the recombination rate for each interval and estimated as (recombination frequency)/(total length of region), where recombination frequency = 100 × (total number of recombinants)/(number of plants) (Lien et al. 2000).
Vector construction and assay of transgenic plants
The construct pCUbi1390-△FAD2 (inserting ubiquitin promoter and a FAD2 intron into pCAMBIA1390) was used as an RNA interference (RNAi) vector (Stoutjesdijk et al. 2002; Li et al. 2013). Both antisense and sense versions of a specific 468-bp fragment from the coding region of open reading frame 3 (ORF3) were amplified (primer pairs TPR-RNAi-A and TPR-RNAi-S; Supplemental Material, Table S1) and successively inserted into pCUbi1390-△FAD2, to form the RNAi construct pUbi-dsRNAiTPR, which was then transformed into parents and heterozygote S7ai/S7cp. The transformation was conducted according to a published method (Hiei et al. 1994).
A fragment containing the whole specific 468-bp fragment and part vector sequence were amplified by using primers RNAi-A (607 bp) and RNAi-S (851 bp) for assay of transgenic positive plants. For background analysis, the transgenic plants were tested with PCR aiming to amplify the S7-containing region with primers TI15 and TI53 (Table S1).
A 2856-bp complementary DNA (cDNA) fragment containing the entire ORF3 coding region and a 1719-bp upstream genomic region were amplified (primer pairs TPRai and TPRai-P) from Ingra; meanwhile, a 2859-bp cDNA fragment and a 1610-bp upstream genomic region were amplified (primer pairs TPRcp and TPRcp-P) from Cpslo17. The PCR products from Ingra and Cpslo17 were inserted into the binary vector pCAMBIA1305, respectively, and then transformed into Cpslo17 and Ingra accordingly. Two primer pairs (P1-F and P2; P3 and P1-R) were used for assay of transgenic positive plants (Table S1).
All transgenic plants were grown either in a nursery in the summer growing season in Beijing or in a greenhouse in winter.
RT-PCR analysis
Total RNA was extracted from the leaf, stem, and root at the seedling stage or from the pistil, lemma, palea, stamen, and panicle at the mature stage using an RNA Prep Pure Plant kit (Tiangen, Beijing) and then reverse transcribed using a SuperScript II kit (TaKaRa). Real-time PCR was performed using a SYBR Premix Ex Taq kit (TaKaRa) on an ABI Prism 7900 Real-Time PCR System. The 2-△△CT method was used to analyze relative changes in gene expression (Livak and Schmittgen 2001). Primers for ORF3 and other two candidate genes (ORF15 and ORF16) were named as 27180-3, 27310-3, and 27320-1. The rice ubiquitin gene (Os03g0234200) was used as a reference in the experiment (primer pair Ubq) (Table S1).
Bioinformatics analysis
Gene prediction was performed using the Rice Genome Annotation Project database (RGAP; http://rice.plantbiology.msu.edu/). Homologous sequences of ORF3 were identified using the Blastp search program of the National Center for Biotechnology Information (NCBI; http://www.ncbi.nlm.nih.gov/). Molecular phylogenetic analysis was constructed by maximum likelihood method using MEGA6 (Tamura et al. 2013). Multiple sequence alignments were conducted with BioEdit software. Prediction of the 3D structure was carried out using phyre2 (http://www.sbg.bio.ic.ac.uk/phyre2/).
Transient expression analysis in Nicotiana benthamiana
The coding sequence of ORF3 from Ingra was amplified and fused to the N terminus of GFP under control of the CaMV 35S promoter in the transient expression vector pCAMBIA1305-GFP, referred to as ORF3-GFP. The cDNA fragment was PCR amplified by the corresponding primer pair (TPR-GFP) (Table S1). Transient expression construct was introduced into the Agrobacterium strain EHA105 and then used to infiltrate N. benthamiana leaves as described previously (Waadt and Kudla 2008). N. benthamiana protoplasts were isolated using the same method as Arabidopsis (Park et al. 2005). Confocal imaging analysis was performed using a Leica TCS SP5 laser scanning confocal microscope.
Data availability
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.
Results
Cytological observation of pollen and embryo sacs in parents and F1 hybrids
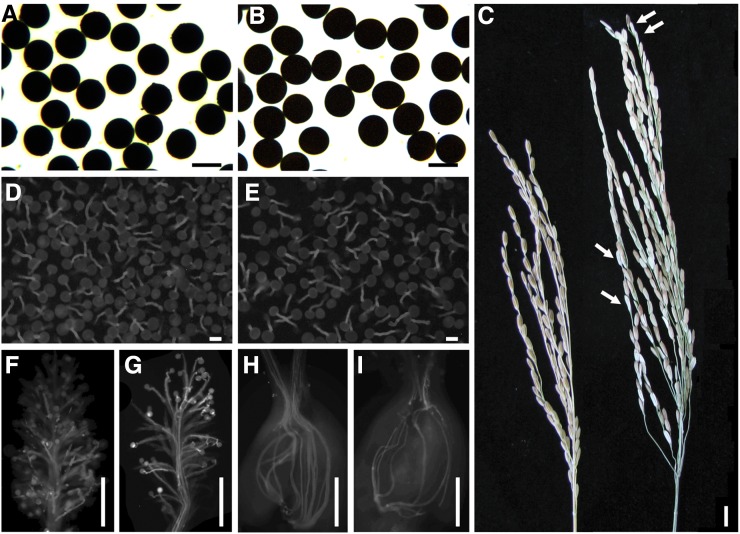
Pollen grains of the three parents and their F1’s were plump and stainable by I2-KI, suggesting pollen fertility of all genotypes was normal (Figure 1, A and B and Table 1). However, spikelet fertility of hybrid F1’s (Ingra/IR36 and Ingra/Cpslo17) showed typically semisterility, compare 49.2 ± 0.1 and 59.6 ± 1.1% of seed setting rates, respectively, against with normal spikelet fertility of their parents (≥80% of seed setting rates, Figure 1C and Table 1).
Figure 1.
Phenotype of Ingra and Ingra/Cpslo17 F1. Pollen I2-KI staining of Ingra (A) and F1 (B). Spikelet of Ingra (left) and F1 (right), the arrow indicates the shriveled grain (C). In vitro pollen germination (D and E), germination of pollen on stigma (F and G), and pollen tube elongation in ovary (H and I) showed no difference between Ingra (D, F, and H) and F1 (E, G, and I). Bars: (A, B, D, and E) 50 μm; (C) 1 cm; and (F–I) 500 μm.
Table 1. The pollen and spikelet fertility of three parents and their F1 hybrids.
| Parents and crosses | Fertility of pollen (%) | Fertility of spikelet (%) |
|---|---|---|
| Ingra | 94.2 ± 2.0 | 95.7 ± 0.8 |
| IR36 | 94.9 ± 2.0 | 84.8 ± 5.2 |
| Cpslo17 | 94.8 ± 1.4 | 86.1 ± 6.2 |
| Ingra/IR36 | 99.1 ± 0.8 | 49.2 ± 0.1*** |
| IR36/Ingra | 88.2 ± 3.3 | 46.8 ± 0.0*** |
| Ingra/Cpslo17 | 89.2 ± 3.5 | 59.6 ± 1.1*** |
| Cpslo17/Ingra | 97.4 ± 0.6 | 54.3 ± 0.4*** |
| Ingra/Nipponbare | 80.8 ± 0.8 | 74.4 ± 0.6 |
| Nipponbare/IR36 | 66.8 ± 1.6 | 50.9 ± 6.7 |
| Ingra/9311 | 99.7 ± 0.3 | 50.2 ± 2.3 |
| 9311/Cpslo17 | 97.5 ± 1.7 | 84.1 ± 6.6 |
| Ingra/Dular | 98.4 ± 1.6 | 87.1 ± 2.0 |
| Cpslo17/Dular | 97.6 ± 0.8 | 86.7 ± 7.6 |
| IR36/Dular | 97.9 ± 0.5 | 92.3 ± 2.4 |
| Ingra/IR24 | 97.6 ± 1.2 | 44.4 ± 4.2 |
| Cpslo17/IR24 | 96.4 ± 1.4 | 92.4 ± 2.7 |
| IR36/IR24 | 95.1 ± 1.3 | 89.9 ± 2.7 |
| Ingra/N22 | 99.0 ± 0.7 | 91.9 ± 1.2 |
| Cpslo17/N22 | 99.1 ± 0.3 | 87.8 ± 6.8 |
| N22/IR36 | 73.4 ± 3.5 | 83.1 ± 3.3 |
Statistically significant difference with respect to their parents; P < 0.001.
To deeply investigate cytological mechanism of hybrid sterility in F1 hybrids, we examined the in vitro germination of pollen grains from parents and their F1’s, which showed that the pollen grains from all plants could germinate efficiently (Figure 1, D and E and Table 2). Furthermore, no distinction was observed between F1’s and their parents when examining the number of pollen grains that adhered to stigmata and pollen tube elongation (Figure 1, F–I). However, the fertilization rate of ovaries measured at 2 days after flowering was lower in F1 hybrids (Table 2). In addition, the spikelet fertility of F1 hybrids was not restored to normal levels after hand pollination with pollen from each parent (Table 2). It suggested that the sterility of F1 hybrids might be mainly due to defects in the female reproductive organs rather than pollen sterility or the frustration of fertilization.
Table 2. The fertility-related traits of three parents and their F1 hybrids.
| Fertility-related traits (%) | Ingra | Cpslo17 | IR36 | Ingra/IR36 | Ingra/Cpslo17 |
|---|---|---|---|---|---|
| In vitro pollen germination rate | 77.5 ± 2.1 | 77.8 ± 1.8 | 79.6 ± 0.1 | 77.5 ± 1.7 | 88.1 ± 1.1 |
| Normal embryo sac | 95.1 ± 1.9 | 85.3 ± 2.4 | 81.4 ± 0.7 | 47.8 ± 5.1 | 50.8 ± 4.4 |
| Fertilized ovaries | 89.4 ± 4.1 | 82.6 ± 1.6 | 92.4 ± 1.6 | 48.2 ± 2.5 | 55.6 ± 4.3 |
| Open seed-setting rate | 95.7 ± 0.8 | 86.1 ± 6.2 | 86.6 ± 4.7 | 49.2 ± 2.2 | 59.6 ± 8.6 |
| Supplementary pollination with parents | — | — | — | 44.3 ± 7.5 | 49.6 ± 4.1 |
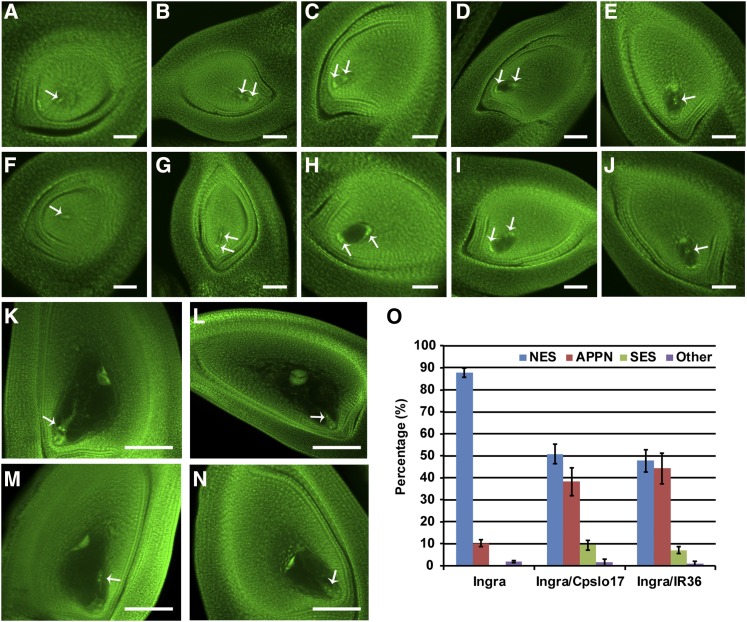
Observation of the embryo sacs from mononucleate stage to mature embryo sac stage showed that abnormal embryo sacs occurred at the eight-nucleate embryo sac developing stage, of which two polar nuclei located in the central cavity with horizontal arrangement instead of vertical one (Figure 2, A–J). There were mainly two different types of abnormalities observed in the mature embryo sacs of the F1 hybrids, including embryo sacs with abnormal polar nuclei positioning and smaller embryo sacs. In the first type, the polar nuclei located either above the egg apparatus with vertical arrangement or near the wall of embryo sac (Figure 2, K–M). In the second type, embryo sacs were smaller, in which abnormal positioning of polar nuclei was found occasionally (Figure 2N). Analysis of histological sections of mature embryo sacs revealed that frequency of abnormal embryo sacs in F1 hybrids was significantly higher than that in parental controls (Figure 2O). Additionally, embryo sac fertility of Ingra/Cpslo17 and Ingra/IR36 F1 corresponded to their spikelet fertility separately (Table 2).
Figure 2.
Observation of embryo sac development. (A and F) Mononucleate embryo sac stage. (B and G) Binucleate embryo sac stage. (C and H) Tetranucleate embryo sac stage. (D and I) Eight-nucleate embryo sac stage. (E and J) Eight-nucleate embryo sac developing stage. (A–E) Ingra. (F–J) Ingra/Cpslo17 F1. (K–N) Embryo sac matured stage, (K) normal embryo sac; (L and M) abnormal positioning of polar nuclei; and (N) small embryo sac. The arrow indicates the polar nuclei. (O) Statistics of different types of embryo sac in mature embryo sac. NES, normal embryo sac; APPN, abnormal positioning of polar nuclei; other, other abnormal embryo sacs; SES, small embryo sac. Bars: (A–J) 50 μm and (K–N) 100 μm.
Taken together, these results suggested that the partial abortive embryo sac caused hybrid sterility. Meanwhile, the failure of normal fertilization in hybrids was mainly caused by abnormal development from the eight-nucleate embryo sac developing stage, which totally induced the occurrence of smaller-sized embryo sacs and embryo sacs with abnormal positioning of polar nuclei, consequently.
Genetic mechanism and neutral allele identification of S7
A framework linkage map was constructed based on a population of 272 F1 plants derived from a three-way cross, Ingra/IR36//Cpslo17, where the average pollen fertility was 90.4 ± 11.9% but the spikelet fertility showed significant bimodal distribution with an apparent valley between ∼70 and 80% (Figure S1 and File S1). The locus showed a tight linkage with Rc (Sweeney et al. 2006) on chromosome 7 (Figure S2 and File S1), which was demonstrated by the fact that nearly all hybrids F2 of both Ingra/Cpslo17 and Ingra/IR36 showed red pericarp inherited from their female parent Ingra (Figure S3). These results confirmed that the locus found in our study is the S7 reported by Yanagihara et al. (1992).
Reciprocal crosses (Ingra/Cpslo17 and Cpslo17; and Ingra/IR36 and IR36) were conducted separately to reveal the genetic mechanism of S7. The segregation of genotypes in progeny was not fit well to Mendel’s law when pollinating Ingra/Cpslo17 and Ingra/IR36 with Cpslo17 and IR36 pollen grains, respectively, of which the number of individuals with homozygote genotype obviously decreased. Since the male and female gametes could be affected differently, two parameters, km and kf, were estimated for TRD. The parameters km of S7cp and S7i alleles, transmitted efficiently through male gametes, were 0.49 and 0.48, respectively. Nevertheless, female gametes were significantly blocked, and the parameters kf of S7cp and S7i alleles were 0.97 and 0.94, respectively. These results indicated the S7ai allele exhibited absolutely preferential transmission by promoting the elimination of S7cp and S7i to their progeny, mostly through the female gamete (Table 3).
Table 3. Genetic analysis of S7.
| Progeny genotypes | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| Female genotype | Male genotype | S7ai/S7ai | S7ai/S7cp | S7cp/S7cp | S7ai/S7i | S7i/S7i | P | TRD | |
| Reciprocal crosses | S7ai/S7cp | S7cp/S7cp | — | 10 | 309 | — | — | 6.6e-63 | kf = 0.97 |
| S7cp/S7cp | S7ai/S7cp | — | 209 | 198 | — | — | 0.59 | km = 0.49 | |
| S7ai/S7i | S7i/S7i | — | — | — | 19 | 293 | 2.9e-54 | kf = 0.94 | |
| S7i/S7i | S7ai/S7i | — | — | — | 202 | 187 | 0.45 | km = 0.48 | |
| Ingra/Cpslo17- ORF3 RNAi | 27-6 | 30 | 50 | 15 | — | — | 0.08 | ||
| 18-5 | 9 | 20 | 5 | — | — | 0.37 | |||
| 13-3 | 33 | 56 | 22 | — | — | 0.33 | |||
P-value obtained using the chi-squared test under the hypothesis of Mendelian segregation.
Based on the model of allelic interaction (Ikehashi and Araki 1984), for three given varieties, A, B, and N, if a hybrid A/B shows gamete abortion, but N/A and N/B do not, the variety N possesses a neutral allele at this locus. To identify the neutral allele of S7, 13 crosses were constructed between the three parents and cultivars including indica and japonica. While the typical sterility was shown in both Ingra/IR36 and Ingra/Cpslo17 F1 hybrids, spikelet fertility was normal when Dular and N22 were crossed with Ingra, IR36, and Cpslo17, which suggested that Dular and N22 probably possess the neutral allele, S7n, at the S7 locus (Table 1).
Fine mapping of S7
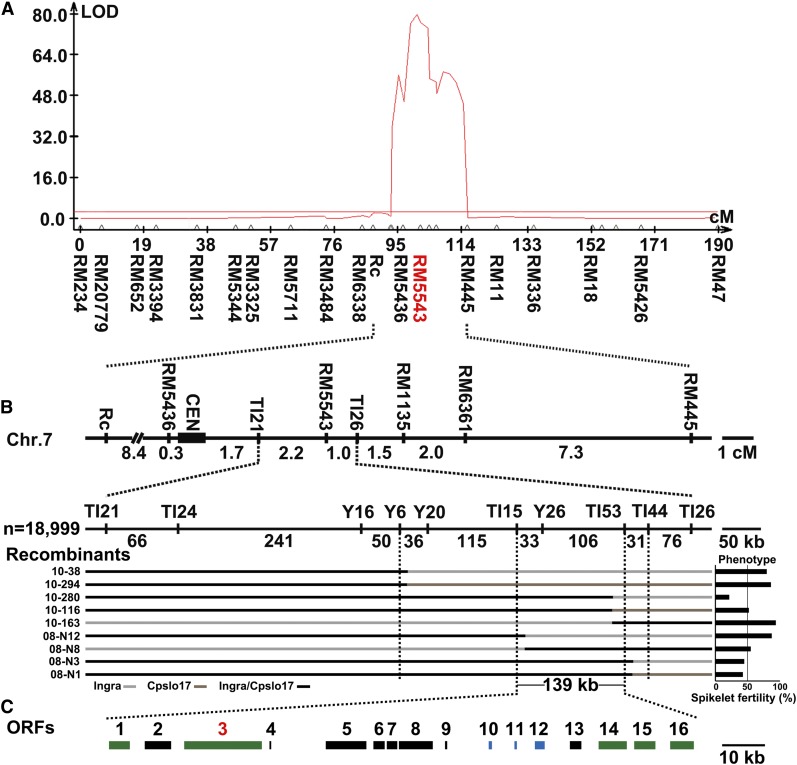
A single, significant QTL was identified on chromosome 7 from the three-way cross Ingra/IR36//Cpslo17 F1 population, suggesting hybrid embryo sac sterility in Ingra/Cpslo17 and Ingra/IR36 was mainly controlled by S7 (Figure 3A). The peak of QTL appeared at molecular marker RM5543 where the LOD score was 80.0. S7 was flanked by Rc and RM445 including the centromere.
Figure 3.
Fine mapping of S7. (A) QTL analysis of S7. (B) The physical map of S7. S7-containing region determined by Ingra/Cpslo17 F2:3 (partial key recombinants were shown from 10–38 to 10–163) and Ingra/IR36//Cpslo17 F2 (partial key recombinants were shown from 08-N12 to 08-N1) population. (C) The overlapped 139 kb included 16 ORFs, of which 8 encode retrotransposon proteins (black boxes), three encode expressed or hypothetical proteins (blue boxes), and the other five have functional annotations (green boxes).
Two populations were used to narrow down the genomic region containing the S7 locus. We initially chose plants carrying S7ai/S7cp between markers Rc and RM445 with a seed setting rate <55% from Ingra/IR36//Cpslo17 F1 and Ingra/Cpslo17 F2 to generate high-resolution mapping populations. Additional markers, including insertion–deletion (InDel) and derived cleaved amplified polymorphic sequences (dCAPSs) were developed in the Rc–RM445 interval. By analyzing recombinants from 12,000 Ingra/Cpslo17 F2:3 plants, the S7 locus was delimited to the interval flanked by Y6 and TI53 with two recombinant events between Y6 and S7 and five between TI53 and S7. Meanwhile, the results from 6999 Ingra/IR36//Cpslo17 F2 populations showed that S7 was delimited to the interval between TH15 and TI44 with three recombinant events on either end (Figure 3B and File S1). The overlapping fragment of 139 kb in the two mapping results included eight ORFs without retrotransposons (RTs) (Figure 3C and Table S2).
Recombination rate
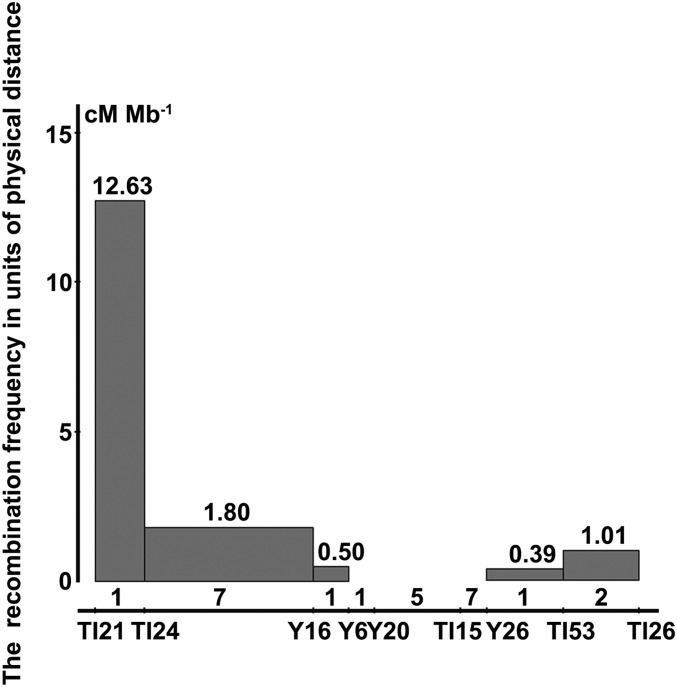
A significant repression of recombination was observed across the primary mapping region of 754 kb (TI21–TI26, Figure 3B) in the centromeric region. The evaluation of 12,000 plants from Ingra/Cpslo17 F2:3 for recombination allowed us to identify recombination frequency in units of physical distance across this region. The most recombinogenic interval was a 66-kb region on the left defined by markers TI21 and TI24. Within this interval, the recombination rate was 12.63 cM/Mb−1, which was significantly higher than that of the 241-kb interval just downstream (TI24 to Y16) and the following 50 kb (Y16 to Y6). A similar case occurred on the right but to a lesser degree, which showed the interval between Y26 and TI53 had a lower recombination rate compared with its downstream 1.01 cM/Mb−1. Besides, the 184-kb interval from Y6 to Y26 had no detectable recombination (Figure 4 and File S1).
Figure 4.
Evaluation of recombination rate across the primary mapping region (754 kb). The x-axis is positioned along the markers used in fine mapping, and the y-axis shows the number of recombination rate, that is the recombination frequency in units of physical distance (cM/Mb−1). The number of retrotransposons is shown above the markers.
While the whole region is rich in RTs, we analyzed the number of RTs in each of the divided regions according to the calculation of recombination rate. Interestingly, in the regions where recombination rate drops sharply, a surge of RTs was also found (Figure 4). Considering the location in the centromere region, it was likely that fewer occurrences of recombination events in Ingra/Cpslo17 F2:3 populations were mainly caused by aggregation of RTs in the target region.
Analysis of candidate genes
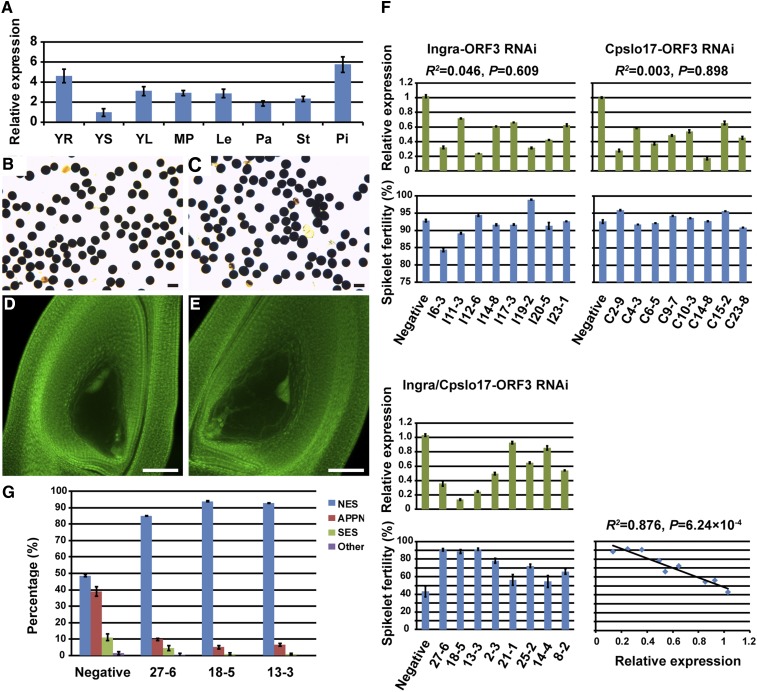
To define candidate gene for S7, we first sequenced the genome of five ORFs (ORF1, 3, and 14–16), which had functional annotations from varieties including parents, Dular (N22), and Ketan Nangka (K.N., possessing the S7kn allele) (Yanagihara et al. 1992). Since amino acid sequences of ORF1 and ORF14 had shown no obvious difference among those varieties, we excluded ORF1 and ORF14 as candidate genes (Figure S4 and File S2). Besides, in contrast to ORF15 and ORF16, which were predominantly expressed in mature panicles, ORF3 was mostly expressed in the pistil (Figure 5A and Figure S5).
Figure 5.
Functional analysis of ORF3. (A) Expression pattern of ORF3. (B–E) Cytological observation of RNAi transgenic plants: negative control (B and D); positive (C and E). (F) Regression analysis of relative expression of ORF3 and spikelet fertility in parent and Ingra/Cpslo17 F1 RNAi transgenic plants. (G) Percentage of different types observed in mature embryo sacs of Ingra/Cpslo17-ORF3 RNAi representative and negative plants. APPN, abnormal positioning of polar nuclei; Le, lemma; MP, mature panicle; NES, normal embryo sac; other, other abnormal embryo sacs; Pa, palea; Pi, pistil; SES, small embryo sac; St, stamen; YL, young leaf; YR, young root; YS, young stem. Bars, 50 μm.
The genomic sequence of ORF3 (LOC_Os07g27180) spans 17,504 bp and contains 19 introns and 20 exons. It encodes a protein of 898 aa with a conserved TPR domain. Compared with others, Ingra showed more specific amino acid sites. Moreover, a variant amino acid was detected at amino acid 119 in Dular and N22 that has a Met (M) instead of a Leu (L) in Ingra, Cpslo17, and K.N. (Figure S6). Based on higher expression in mature pistils and unique differences in amino acid sequence, ORF3 was selected as the candidate gene for S7 tentatively.
S7 encodes a TPR-containing protein
To investigate function of ORF3, we knocked down ORF3 in parents and heterozygote S7ai/S7cp by RNAi. A total of 26 Ingra-ORF3 RNAi, 33 Cpslo17-ORF3 RNAi, and 53 Ingra/Cpslo17-ORF3 RNAi transgenic-positive T0 plants were obtained by background detection and transgenic PCR assay (Figure S7 and Figure S8). The pollen grains in all ORF3 RNAi plants showed no obvious abortion compared with those in transgenic-negative plants, while embryo sacs with abnormal positioning of polar nuclei and small embryo sacs were significantly reduced in Ingra/Cpslo17-ORF3 RNAi plants (Figure 5, B–E). ORF3 RNAi transgenic positive plants had no effect on spikelet fertility of both Ingra and Cpslo17. However, significantly restored spikelet fertility was observed in Ingra/Cpslo17-ORF3 RNAi-positive plants (Table 4).
Table 4. Spikelet fertility of transgenic T0 plants.
| Receptor | Types | No. of plants | Spikelet fertility (%) |
|---|---|---|---|
| Ingra-ORF3 RNAi | Positive | 26 | 92.03 |
| Negative | 12 | 92.37 | |
| t | 2.03 | ||
| P | 0.84 | ||
| Cpslo17-ORF3 RNAi | Positive | 33 | 93.54 |
| Negative | 13 | 92.17 | |
| t | 2.12 | ||
| P | 0.15 | ||
| Ingra/Cpslo17-ORF3 RNAi | Positive | 53 | 77.00 |
| Negative | 19 | 48.04 | |
| t | 2.00 | ||
| P | 0.00 | ||
| Cpslo17-S7ai | Positive | 11 | 49.00 |
| Negative | 6 | 82.19 | |
| t | 2.18 | ||
| P | 0.00 | ||
| Ingra-S7cp | Positive | 7 | 90.03 |
| Negative | 12 | 93.56 | |
| t | 2.11 | ||
| P | 0.08 |
In order to verify the correlation between relative expression of ORF3 and spikelet fertility, eight families with different spikelet fertility levels from Ingra-ORF3 RNAi, Cpslo17-ORF3 RNAi, and Ingra/Cpslo17-ORF3 RNAi plants were randomly selected, respectively. Gene expression in mature pistils was examined by RT-RCR. In parental transgenic plants, correlation between relative expression of ORF3 and spikelet fertility was not obvious. However, the extent of downregulation was consistent with the restored level of spikelet fertility in Ingra/Cpslo17-ORF3 RNAi-positive plants. Strong interference plants (such as 27-6, 18-5, and 13-3) exhibited a high fertility rate, whereas weak interference plants (such as 21-1 and 14-4) were associated with a low fertility rate (Figure 5F).
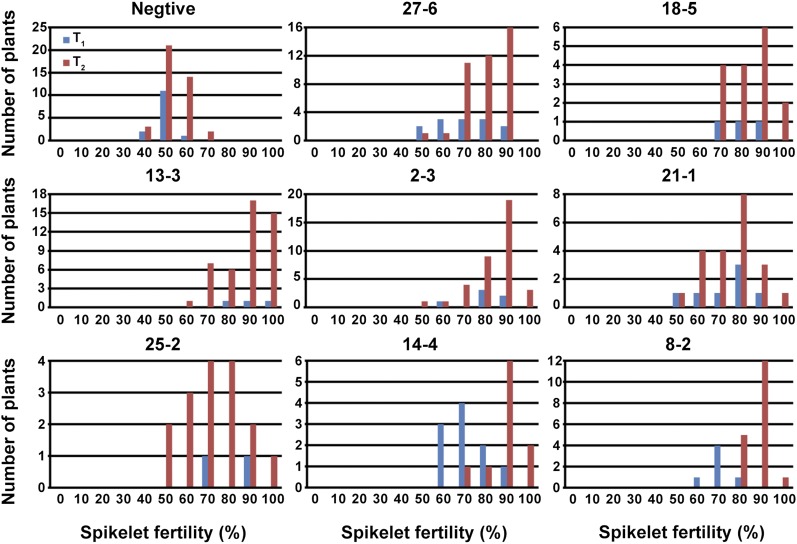
To test the hypothesis that the restored spikelet fertility of the ORF3 RNAi plants might be mainly due to normal development of the embryo sac, we selected three strong interference plants (27-6, 18-5, and 13-3) as representative ORF3 RNAi plants for further analysis. The rate of embryo sac abortion in representative plants ranged from 6.13 to 15.04%, which was significantly lower than 51.46% detected in the negative plants. Correspondingly, the frequency of normal embryo sacs in the representative plants increased substantially (Figure 5G). These data were in accordance with the spikelet fertility in the RNAi positive and negative plants. The offspring of the eight RNAi transgenic positive plants showed a similar phenotype of restored spikelet fertility from the T1 to T2 generation (Figure 6 and Table S3). Additionally, segregation ratio in T2 generation of those three representative ORF3 RNAi families conformed to Mendel’s law (Table 3). These results demonstrated that ORF3 RNAi could restore spikelet fertility of heterozygote S7ai/S7cp and eliminate absolutely the preferential transmission of S7ai.
Figure 6.
Distribution of spikelet fertility of Ingra/Cpslo17-ORF3 RNAi from T1 to T2 generation.
In addition, complementation tests were performed to confirm the function of ORF3. The strategy that we adopted was to transform the S7ai allele into Cpslo17 and the S7cp allele into Ingra, respectively. Through PCR assay, a series of transgenic-positive T0 plants was obtained (Figure S9). Examination of the spikelet fertility showed that there was no statistically significant difference between the transgenic-positive and -negative plants of Ingra-S7cp. By contrast, transgenic-positive and -negative plants of the Cpslo17-S7ai showed a highly significant difference in spikelet fertility; the average spikelet fertility of the positive plants (49.00%) was much lower than that of the negative plants (82.19%) (Table 4). Considering the S7ai allele exhibited absolute preferential transmission by promoting the elimination of S7cp, we thus concluded that ORF3 is the candidate gene.
Expression and subcellular location of ORF3
Phylogenetic tree analysis showed that ORF3 exists in not only Asian rice (O. sativa L.) but also wild rice. Cpslo17 is closely related to the indica type and distanced from Ingra, confirming the similarity with IR36, which could not transmit their female gamete to their progeny in hybrid F1’s crossed with Ingra. Homologous proteins of ORF3 were identified in other plants, all of which belong to the grass family (Figure S10). Furthermore, protein sequence alignment suggested that ORF3 exhibited 76.0–79.7% identity with its homologous proteins, indicating that the ORF3 is specific to monocots (Figure S11).
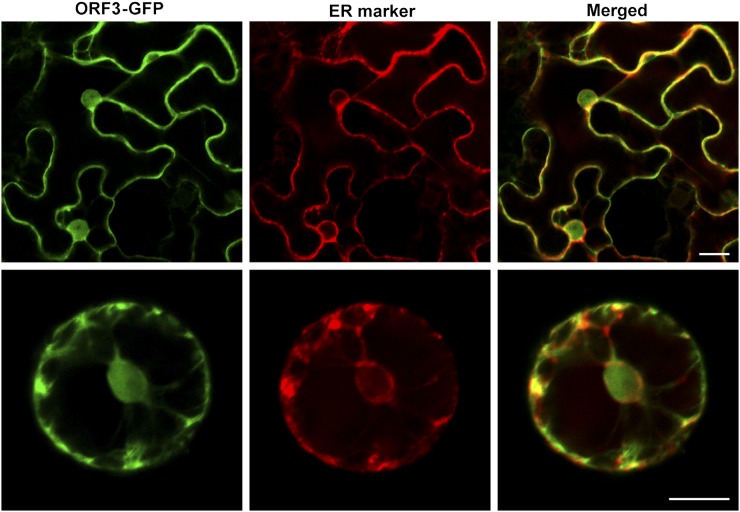
To determine experimentally subcellular localization, a fusion protein, ORF3-GFP, was generated and expressed transiently under the control of the 35S promoter in tobacco (N. benthamiana) mesophyll protoplasts. As shown in Figure 7, ORF3-GFP was detected in the ER and nuclei.
Figure 7.
Subcellular localization of ORF3 protein in tobacco mesophyll protoplasts. Bars, 20 μm.
Discussion
Rice is one of the main crops providing staple food for more than half the global population. Utilization of strong heterosis in intersubspecific crosses of rice has long been difficult due to semisterility of panicles in the hybrids between indica and japonica cultivars. One of the genes involved in this phenomenon is S7, which has been isolated here through a positional cloning approach. We demonstrate that a TPR-containing protein plays a significant role in hybrid female sterility, which is particularly important for understanding the molecular mechanism of intersubspecific hybrid sterility in rice.
Evidence from the current study suggests that the function of S7 is centered on functional female gametophyte formation. Abnormal female gametophyte of hybrid F1 appears at the eight-nucleate embryo sac developing stage, when polar nuclei start their migration to the upward side of the egg apparatus and the embryo sac begins to enlarge (Figure 2). In addition, the abnormal embryo sac caused by S7 led to the semisterility of spikelets directly (Table 2), which was consistent with a previous report that most of the smaller embryo sacs and abnormal polar nuclei positioning embryo sacs could not succeed in fertilization, even with a normal female germ unit (Zeng et al. 2009). It was reported that orientation of polar nuclei always occurred not only during development of female gametophyte but also after fertilization, when one sperm cell nucleus moved toward the central cell (Ding et al. 2009). Observation on the distribution and structural organization of the microtubule during megagametogenesis suggested that some new organizational patterns of microtubules surrounding the central cell might be associated with the probable movement and positioning of the polar nuclei (Xu et al. 2001). Furthermore, an involvement of F-actin in gamete nuclear migration had been suggested that F-actin disorganization in the central cell could disrupt the polarized nuclear location and the presence of intact F-actin cables in the central cell was correlated with successful karyogamy regardless of the central cell nuclear position (Kawashima et al. 2014; Ohnishi et al. 2014; Kawashima and Berger 2015). Therefore, we speculate that some cytoskeleton or signal molecules, involved in the migration of polar nuclei, were disorganized during the development of the embryo sac and led to incorrect guidance of polar nuclei fertilization eventually in both Ingra/IR36 and Ingra/Cpslo17 hybrids.
Analysis of recombination rate revealed that a large number of RTs, especially Ty3-gypsy, were aggregated in the S7-containing region near the centromere (Figure 4). In general, transposable elements (TEs) were found to be enriched in the pericentromeric regions of many plant genomes (Arabidopsis Genome Initiative 2000; International Rice Genome Sequencing Project 2005; Paterson et al. 2009; Schnable et al. 2009; Schmutz et al. 2010; Luo et al. 2012). Genes close to the centromere or retrotransposon clusters are less recombinogenic than other genes in a gene-rich region (Copenhaver et al. 1999; Fu et al. 2001). Significant recombinational repressions were reported in the mapping regions of Rc/rg7.1 (Sweeney et al. 2006) and Ghd7 (Xue et al. 2008). Both of the genes located close to S7, demonstrating that the low frequency of recombination in the S7-containing region may be due to its particular location in the centromeric region on chromosome 7. In plants, self-incompatibility genes were found to be recombinationally suppressed due to their subcentromeric location in Petunia (Coleman and Kao 1992; Entani et al. 1999), Antirrhinum (Ma et al. 2003; Yang et al. 2007), and the presence of repetitive DNA in Nicotiana (Matton et al. 1995). Additionally, the hybrid incompatibility genes Lhr, Zhr, and OdsH in Drosophila, were all mapped to recombinationally suppressed pericentric and heterochromatic regions with reduced or undetectable levels of recombination (Sawamura et al. 1993; Brideau et al. 2006; Bayes and Malik 2009). Consequently, it is still necessary to validate whether the suppression of recombination plays a role in species conservation.
The TPR domain is one of the most frequently observed amino acid motifs in nature, which can be found in numerous proteins. TPR-containing proteins always serve as interaction modules and multiprotein complex mediators (Blatch and Lassle 1999; Zeytuni et al. 2011; Zeytuni and Zarivach 2012; Shin et al. 2014) and regulate diverse biological processes, including reproductive development. In rice, mutation in OsAPC6, one of the TPR-containing anaphase-promoting complex/cyclosome (APC/C) subunits, caused a reduced number or complete absent of polar nuclei (Kumar et al. 2010; Awasthi et al. 2012). Therefore, protein–protein interaction may provide important clues to reveal the molecular mechanism of S7. In our study, downregulated expression of ORF3 could restore spikelet fertility in the hybrid F1 Ingra/Cpslo17. However, there was no effect in either Ingra-ORF3 RNAi or Cpslo17-ORF3 RNAi plants (Figure 5 and Table 4). These results suggest that S7 may not be essential for growth, development, or reproduction, and hybrid sterility of Ingra/Cpslo17 is probably controlled by allelic interaction between S7ai and S7cp. Therefore, weak allelic interaction in Ingra/Cpslo17-ORF3 RNAi plants could overcome the hybrid sterility. Although transformants Cpslo17-S7ai and Ingra-S7cp have S7ai and S7cp alleles, simultaneously, the sterility occurs only in S7ai/S7ai-S7cp (Table 4). Combined with the results of TRD analysis (Table 3), the S7ai allele exhibits stronger function during female gametes transferring in heterozygote (S7ai/S7cp) plants. Due to the fact that the F1 hybrids between three parents (Ingra, IR36, and Cpslo17) and wide-compatibility varieties (WCVs) (Dular and N22) showed normal spikelet fertility (Table 1), Dular and N22 were assumed to carry the neutral allele S7n at the S7 locus. To confirm the function of S7n, we sequenced the S7 region in three parents and WCVs and found that a SNP (C/A) caused an amino acid substitution (Leu-119 in parents, Met-119 in Dular and N22) (Figure S6 and Table S4). Additionally, a total of 47 varieties including indica, japonica, and wild rice were compared with the genomic sequence of the S7 region, the result of which indicated that the nucleotide C exists in most varieties (Table S4). However, Leu-119 is conserved not only in various rice species, but also in other plants (Figure S10). Moreover, predicted 3D structures of ORF3 from Ingra, Cpslo17, and Dular indicated that amino acid sequence differences may induce their changes in spatial structure directly (Figure S12). Therefore, we speculate that during the formation of female gametophytes in heterozygotes S7ai/S7cp and S7ai/S7i, gametes carrying S7cp and S7i could not resist the effect of S7ai from sporophyte, thus leading to embryo sac abortion. However, the S7ai gametes could resolve such function of S7cp and S7i, exhibiting preferential transmission. Additionally, substitution of Met-119 in S7n may cause loss of function of protein–protein interactions in Dular and N22, which results in the failure to produce sterile offspring when crossed with either indica or japonica (Figure S13). Although how such likely nonfunction is related to hybrid fertility remains to be characterized, the discovery and molecular analysis of S7n in this study probably can provide functional markers for WCG germplasm screening to solve, at least partially, hybrid embryo sac sterility in rice breeding.
Acknowledgments
This research was supported by the National Transgenic Science and Technology Program (2013ZX08001004-002 and 2013ZX08009003-003), the Chinese National High Technology Research and Development Program (“863” 2014AA10A604), the Key Research and Development Program of Jiangsu Province (BE2015363), and grants from the Ministry of Agriculture Key Laboratory of the middle and lower reaches of the Yangtze River japonica rice biology and genetic breeding.
Footnotes
Communicating editor: J. A. Birchler
Supplemental material is available online at www.genetics.org/lookup/suppl/doi:10.1534/genetics.115.183848/-/DC1.
Literature Cited
- Arabidopsis Genome Initiative , 2000. Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408: 796–815. [DOI] [PubMed] [Google Scholar]
- Awasthi A., Paul P., Kumar S., Verma S. K., Prasad R., et al. , 2012. Abnormal endosperm development causes female sterility in rice insertional mutant OsAPC6. Plant Sci. 183: 167–174. [DOI] [PubMed] [Google Scholar]
- Baack E., Melo M. C., Rieseberg L. H., Ortiz-Barrientos D., 2015. The origins of reproductive isolation in plants. New Phytol. 207: 968–984. [DOI] [PubMed] [Google Scholar]
- Barbash D. A., Siino D. F., Tarone A. M., Roote J., 2003. A rapidly evolving MYB-related protein causes species isolation in Drosophila. Proc. Natl. Acad. Sci. USA 100: 5302–5307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayes J. J., Malik H. S., 2009. Altered heterochromatin binding by a hybrid sterility protein in Drosophila sibling species. Science 326: 1538–1541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bikard D., Patel D., Le Mette C., Giorgi V., Camilleri C., et al. , 2009. Divergent evolution of duplicate genes leads to genetic incompatibilities within A. thaliana. Science 323: 623–626. [DOI] [PubMed] [Google Scholar]
- Blatch G. L., Lassle M., 1999. The tetratricopeptide repeat: a structural motif mediating protein-protein interactions. BioEssays 21: 932–939. [DOI] [PubMed] [Google Scholar]
- Brideau N. J., Flores H. A., Wang J., Maheshwari S., Wang X., et al. , 2006. Two Dobzhansky-Muller genes interact to cause hybrid lethality in Drosophila. Science 314: 1292–1295. [DOI] [PubMed] [Google Scholar]
- Chen J., Jiang L., Wang C. M., Ikehashi H., Zhai H. Q., et al. , 2006. Mapping of loci for pollen sterility of indica-japonica hybrids in rice (Oryza sativa L.). Acta Agron. Sin. 32: 515–521. [Google Scholar]
- Chen J. J., Ding J. H., Ouyang Y. D., Du H. Y., Yang J. Y., et al. , 2008. A triallelic system of S5 is a major regulator of the reproductive barrier and compatibility of indica-japonica hybrids in rice. Proc. Natl. Acad. Sci. USA 105: 11436–11441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen M. J., Zhao Z. G., Jiang L., Wan J. M., 2012. A new gene controlling hybrid sterility in rice (Oryza sativa L.). Euphytica 184: 15–22. [Google Scholar]
- Chen S., Luo Z. L., Zhang D. X., 2014. Pre- and post-zygotic reproductive isolation between co-occurring Mussaenda pubescens var. alba and M. shikokiana (Rubiaceae). J. Integr. Plant Biol. 56: 411–419. [DOI] [PubMed] [Google Scholar]
- Coleman C. E., Kao T., 1992. The flanking regions of two Petunia inflata S alleles are heterogeneous and contain repetitive sequences. Plant Mol. Biol. 18: 725–737. [DOI] [PubMed] [Google Scholar]
- Copenhaver G. P., Nickel K., Kuromori T., Benito M. I., Kaul S., et al. , 1999. Genetic definition and sequence analysis of Arabidopsis centromeres. Science 286: 2468–2474. [DOI] [PubMed] [Google Scholar]
- Coyne J. A., Orr H. A., 2004. Speciation, Sinauer Associates, Sunderland, MA. [Google Scholar]
- Dai X. M., Huang Q. C., Qin G. Y., Li G. P., 2006. Observation on particular embryo sac of rice using confocal microscopy. J. North China Agriculture 21: 26–29. [Google Scholar]
- De Vienne D. M., Refregier G., Hood M. E., Guigue A., Devier B., et al. , 2009. Hybrid sterility and inviability in the parasitic fungal species complex Microbotryum. J. Evol. Biol. 22: 683–698. [DOI] [PubMed] [Google Scholar]
- Ding, J. T., J. H. Shen, W. Li, and H. Yang, 2009 Cytological observation of double fertilization and its duration in Oryza sativa. Bull. Bot. 44: 473–483.
- Dobzhansky T., 1937. Genetics and the Origin of Species, Columbia University Press, New York. [Google Scholar]
- Entani T., Iwano M., Shiba H., Takayama S., Fukui K., et al. , 1999. Centromeric localization of an S-RNase gene in Petunia hybrida Vilm. Theor. Appl. Genet. 99: 391–397. [DOI] [PubMed] [Google Scholar]
- Ferree P. M., Barbash D. A., 2009. Species-specific heterochromatin prevents mitotic chromosome segregation to cause hybrid lethality in Drosophila. PLoS Biol. 7: e1000234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu H. H., Zheng Z. W., Dooner H. K., 2001. Recombination rates between adjacent genic and retrotransposon regions in maize vary by 2 orders of magnitude. Proc. Natl. Acad. Sci. USA 99: 1082–1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grant V., 1981. Plant Speciation, Columbia University Press, New York. [Google Scholar]
- Hiei Y., Ohta S., Komari T., Kumashiro T., 1994. Efficient transformation of rice (Oryza sativa L.) mediated by Agrobacterium and sequence analysis of the boundaries of the T-DNA. Plant J. 6: 271–282. [DOI] [PubMed] [Google Scholar]
- Hughes S., Wilkinson H., Gilbert S. P., Kishida M., Ding S. S., et al. , 2014. The C. elegans TPR containing protein, TRD-1, regulates cell fate choice in the developing germ line and epidermis. PLoS One 9: e114998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikehashi H., Araki H., 1984. Variety screening of compatibility types revealed in F1 fertility of distant cross in rice. Japanese J. Breed. 34: 304–313. [Google Scholar]
- Ikehashi H., Araki H., 1987. Screening and genetic analysis of wide-compatibility in F1 hybrids of distant crosses in rice (Oryza sativa L.). Tech. Bull. Trop. Agric. Res. 23: 1–79. [Google Scholar]
- International Rice Genome Sequencing Project , 2005. The map-based sequence of the rice genome. Nature 436: 793–800. [DOI] [PubMed] [Google Scholar]
- Ji Q., Lu J. F., Chao Q., Zhang Y., Zhang M. J., et al. , 2010. Two sequence alterations, a 136 bp InDel and an A/C polymorphic site, in the S5 locus are associated with spikelet fertility of indica-japonica hybrid in rice. J. Genet. Genomics 37: 57–68. [DOI] [PubMed] [Google Scholar]
- Jing W., Zhang W. W., Jiang L., Chen L. M., Zhai H. Q., et al. , 2007. Two novel loci for pollen sterility in hybrids between the weedy strain Ludao and the japonica variety Akihikari of rice (Oryza sativa L.). Theor. Appl. Genet. 114: 915–925. [DOI] [PubMed] [Google Scholar]
- Kawashima T., Berger F., 2015. The central cell nuclear position at the micropylar end is maintained by the balance of F-actin dynamics, but dispensable for karyogamy in Arabidopsis. Plant Reprod. 28: 103–110. [DOI] [PubMed] [Google Scholar]
- Kawashima T., Maruyama D., Shagiron M., Li J., Hamamura Y., et al. , 2014. Dynamic F-actin movement is essential for fertilization in Arabidopsis thaliana. eLife 3: e04501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koide Y., Onishi K., Nishimoto D., Baruah A. R., Kanazawa A., et al. , 2008. Sex-independent transmission ratio distortion system responsible for reproductive barriers between Asian and African rice species. New Phytol. 179: 888–900. [DOI] [PubMed] [Google Scholar]
- Koide Y., Shinya Y., Ikenaga M., Sawamura N., Matsubara K., et al. , 2012. Complex genetic nature of sex-independent transmission ratio distortion in Asian rice species: the involvement of unlinked modifiers and sex-specific mechanisms. Heredity 108: 242–247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo T., Yamagata Y., Eguchi M., Yoshimura A., 2008. A novel epistatic interaction at two loci causing hybrid male sterility in an inter-subspecific cross of rice (Oryza sativa L.). Genes Genet. Syst. 83: 443–453. [DOI] [PubMed] [Google Scholar]
- Kubo T., Yoshimura A., Kurata N., 2011. Hybrid male sterility in rice is due to epistatic interactions with a pollen killer locus. Genetics 189: 1083–1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar M., Basha O. P., Puri A., Rajpurohit D., Randhawa G. S., et al. , 2010. A candidate gene OsAPC6 of anaphase-promoting complex of rice identified through T-DNA insertion. Funct. Integr. Genomics 10: 349–358. [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Chou J. Y., Cheng L., Chang N. H., Yang S. Y., et al. , 2008. Incompatibility of nuclear and mitochondrial genomes causes hybrid sterility between two yeast species. Cell 135: 1065–1073. [DOI] [PubMed] [Google Scholar]
- Levin D. A., 1978. The origin of isolating mechanisms in flowering plants. Evol. Biol. 11: 185–317. [Google Scholar]
- Li D. T., Chen L. M., Jiang L., Zhu S. S., Zhao Z. G., et al. , 2007. Fine mapping of S32(t), a new gene causing hybrid embryo sac sterility in a Chinese landrance rice (Oryza sativa L.). Theor. Appl. Genet. 114: 515–524. [DOI] [PubMed] [Google Scholar]
- Li H., Jiang L., Youn J. H., Sun W., Cheng Z. J., et al. , 2013. A comprehensive genetic study reveals a crucial role of CYP90D2/D2 in regulating plant architecture in rice (Oryza sativa). New Phytol. 200: 1076–1088. [DOI] [PubMed] [Google Scholar]
- Li J., Liu J., Wang G., Cha J. Y., Li G., et al. , 2015. A chaperone function of NO CATALASE ACTIVITY1 is required to maintain catalase activity and for multiple stress responses in Arabidopsis. Plant Cell 27: 908–925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lien S., Szyda J., Schechinger B., Rappold G., Arnheim N., 2000. Evidence for heterogeneity in recombination in the human pseudoautosomal region: high resolution analysis by sperm typing and radiation-hybrid mapping. Am. J. Hum. Genet. 66: 557–566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Arciga-Reyes L., Zhong S., Alexander L., Hackett R., et al. , 2008. SlTPR1, a tomato tetratricopeptide repeat protein, interacts with the ethylene receptors NR and LeETR1, modulating ethylene and auxin responses and development. J. Exp. Bot. 59: 4271–4287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D., 2001. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods 25: 402–408. [DOI] [PubMed] [Google Scholar]
- Long Y. M., Zhao L. F., Niu B. X., Su J., Wu H., et al. , 2008. Hybrid male sterility in rice controlled by interaction between divergent alleles of two adjacent genes. Proc. Natl. Acad. Sci. USA 105: 18871–18876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo S., Mach J., Abramson B., Ramirez R., Schurr R., et al. , 2012. The cotton centromere contains a Ty3-gypsy-like LTR retroelement. PLoS One 7: e35261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma W. S., Zhou J. L., Lai Z., Zhang Y. S., Xue Y. B., 2003. The self-incompatibilty S locus of Antirrhinum resides in a pericentromeric region. Acta Bot. Sin. 45: 47–52. [Google Scholar]
- Masuda K., Chiyoda T., Sugiyama N., Segura-Cabrera A., Kabe Y., et al. , 2015. LATS1 and LATS2 phosphorylate CDC26 to modulate assembly of the tetratricopeptide repeat subcomplex of APC/C. PLoS One 10: e0118662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matton D. P., Mau S. L., Okamoto S., Clarke A. E., Newbigin E., 1995. The S-locus of Nicotiana alata: genomic organization and sequence analysis of two S-RNase alleles. Plant Mol. Biol. 28: 847–858. [DOI] [PubMed] [Google Scholar]
- Mayr E., 1942. Systematics and the Origin of Species, Columbia University Press, New York. [Google Scholar]
- Mayr E., 1963. Animal Species and Evolution, Harvard University Press, Cambridge, MA. [Google Scholar]
- Mizuta Y., Harushima Y., Kurata N., 2010. Rice pollen hybrid incompatibility caused by reciprocal gene loss of duplicated genes. Proc. Natl. Acad. Sci. USA 107: 20417–20422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohnishi Y., Hoshino R., Okamoto T., 2014. Dynamics of male and female chromatin during karyogamy in rice zygotes. Plant Physiol. 165: 1533–1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Lee D., Lee G. J., Hwang I., 2005. AtRMR1 functions as a cargo receptor for protein trafficking to the protein storage vacuole. J. Cell Biol. 170: 757–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paterson A. H., Bowers J. E., Bruggmann R., Dubchak I., Grimwood J., et al. , 2009. The Sorghum bicolor genome and the diversification of grasses. Nature 457: 551–556. [DOI] [PubMed] [Google Scholar]
- Phadnis N., Orr H. A., 2009. A single gene causes both male sterility and segregation distortion in Drosophila hybrids. Science 323: 376–379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Presgraves D. C., Balagopalan L., Abmayr S. M., Orr H. A., 2003. Adaptive evolution drives divergence of a hybrid inviability gene between two species of Drosophila. Nature 423: 715–719. [DOI] [PubMed] [Google Scholar]
- Sawamura K., Yamamoto M. T., Watanabe T. K., 1993. Hybrid lethal systems in the Drosophila melanogaster species complex. II. The Zygotic hybrid rescue (Zhr) gene of D. melanogaster. Genetics 133: 307–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmutz J., Cannon S. B., Schlueter J., Ma J., Mitros T., et al. , 2010. Genome sequence of the palaeopolyploid soybean. Nature 463: 178–183. [DOI] [PubMed] [Google Scholar]
- Schnable P. S., Ware D., Fulton R. S., Stein J. C., Wei F., et al. , 2009. The B73 maize genome: complexity, diversity, and dynamics. Science 326: 1112–1115. [DOI] [PubMed] [Google Scholar]
- Schreiber D. N., Dresselhaus T., 2003. In vitro pollen germination and transient transformation of Zea mays and other plant species. Plant Mol. Biol. Report. 21: 31–41. [Google Scholar]
- She K. C., Kusano H., Koizumi K., Yamakawa H., Hakata M., et al. , 2010. A novel factor FLOURY ENDOSPERM2 is involved in regulation of rice grain size and starch quality. Plant Cell 22: 3280–3294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shin S. B., Golovkin M., Reddy A. S., 2014. A pollen-specific calmodulin-binding protein, NPG1, interacts with putative pectate lyases. Sci. Rep. 4: 5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoutjesdijk, P. A., S. P. Singh, Q. Liu, C. J. Hurlstone, P. A. Waterhouse et al., 2002 hpRNA-mediated targeting of the Arabidopsis FAD2 gene gives highly efficient and stable silencing. Plant Physiol. 129: 1723–1731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M., McCouch S., 2007. The complex history of the domestication of rice. Ann. Bot. (Lond.) 100: 951–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweeney M., Thomson M., Pfeil B., McCouch S., 2006. Caught red-handed: Rc encodes a basic helix-loop-helix protein conditioning red pericarp in rice. Plant Cell 18: 283–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweigart A. L., Willis J. H., 2012. Molecular evolution and genetics of postzygotic reproductive isolation in plants. F1000 Biol. Rep. 4: 23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura K., Stecher G., Peterson D., Filipski A., Kumar S., 2013. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30: 2725–2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang S., Presgraves D. C., 2009. Evolution of the Drosophila nuclear pore complex results in multiple hybrid incompatibilities. Science 323: 779–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ting C. T., Tsaur S. C., Wu M. L., Wu C. I., 1998. A rapidly evolving homeobox at the site of a hybrid sterility gene. Science 282: 1501–1504. [DOI] [PubMed] [Google Scholar]
- Ting Y., 1957. The origin and evolution of cultivated rice in China. Acta Bot. Sin. 8: 243–260 (In Chinese). [Google Scholar]
- Tranguch S., Cheung-Flynn J., Daikoku T., Prapapanich V., Cox M. B., et al. , 2005. Cochaperone immunophilin FKBP52 is critical to uterine receptivity for embryo implantation. Proc. Natl. Acad. Sci. USA 102: 14326–14331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Kudla J., 2008. In planta visualization of protein interactions using bimolecular fluorescence complementation (BiFC). Cold Spring Harb. Protoc. 2008: t4995. [DOI] [PubMed] [Google Scholar]
- Wan J. M., Ikehashi H., 1995. Identification of a new locus S-16 causing hybrid sterility in native rice varieties (Oryza sativa L.) from Tai-hu lake region and Yunnan province, China. Breed. Sci. 45: 161–170. [Google Scholar]
- Wan J., Yanagihara S., Kato H., Ikehashi H., 1993. Multiple alleles at a new locus causing hybrid sterility between a Korean indica variety and japonica variety in rice. Japanese J. Breed. 43: 507–516. [Google Scholar]
- Wan J., Yamaguchi Y., Kato H., Ikehashi H., 1996. Two new loci for hybrid sterility in cultivated rice (Oryza sativa L.). Theor. Appl. Genet. 92: 183–190. [DOI] [PubMed] [Google Scholar]
- Wan J. M., Ikehashi H., 1996. Evidence for mutational origin of hybrid sterility genes in rice (Oryza sativa L.). Breed. Sci. 46: 167–172. [Google Scholar]
- Wang K. L., Yoshida H., Lurin C., Ecker J. R., 2004. Regulation of ethylene gas biosynthesis by the Arabidopsis ETO1 protein. Nature 428: 945–950. [DOI] [PubMed] [Google Scholar]
- White M. A., Steffy B., Wiltshire T., Payseur B. A., 2011. Genetic dissection of a key reproductive barrier between nascent species of house mice. Genetics 189: 289–304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer A., Lexer C., Cozzolino S., 2009. Evolution of reproductive isolation in plants. Heredity 102: 31–38. [DOI] [PubMed] [Google Scholar]
- Xu S. X., Liu X. D., Zhu H. L., Lu Y. G., 2001. Further studies on microtubule organizational changes during megagametogenesis in rice embryo sac. Acta Bot. Sin. 43: 910–917. [Google Scholar]
- Xue W. Y., Xing Y. Z., Weng X. Y., Zhao Y., Tang W. J., et al. , 2008. Natural variation in Ghd7 is an important regulator of heading date and yield potential in rice. Nat. Genet. 40: 761–767. [DOI] [PubMed] [Google Scholar]
- Yamagata Y., Yamamoto E., Aya K., Win K. T., Doi K., et al. , 2010. Mitochondrial gene in the nuclear genome induces reproductive barrier in rice. Proc. Natl. Acad. Sci. USA 107: 1494–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yanagihara S., Kato H., Ikehashi H., 1992. A new locus for multiple alleles causing hybrid sterility between an Aus variety and Javanica varieties in rice (Oryza sativa L.). Japanese Journal of Breeding 42: 793–801. [Google Scholar]
- Yang J. Y., Zhao X. B., Cheng K., Du H. Y., Ouyang Y. D., et al. , 2012. A killer-protector system regulates both hybrid sterility and segregation distortion in rice. Science 337: 1336–1340. [DOI] [PubMed] [Google Scholar]
- Yang Q. Y., Zhang D. F., Li Q., Cheng Z. K., Xue Y. B., 2007. Heterochromatic and genetic features are consistent with recombination suppression of the self-incompatibility locus in Antirrhinum. Plant J. 51: 140–151. [DOI] [PubMed] [Google Scholar]
- Yang Z., Wolf I. M., Chen H., Periyasamy S., Chen Z., et al. , 2006. FK506-binding protein 52 is essential to uterine reproductive physiology controlled by the progesterone receptor A isoform. Mol. Endocrinol. 20: 2682–2694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng Y. X., Hu C. Y., Lu Y. G., Li J. Q., Liu X. D., 2009. Abnormalities occurred during female gametophyte development result in the diversity of abnormal embryo sacs and lead to the abnormal fertilization in indica/japonica hybrids in rice. J. Integr. Plant Biol. 51: 3–12. [DOI] [PubMed] [Google Scholar]
- Zeytuni N., Zarivach R., 2012. Structural and functional discussion of the tetra-trico-peptide repeat, a protein interaction module. Structure 20: 397–405. [DOI] [PubMed] [Google Scholar]
- Zeytuni N., Ozyamak E., Ben-Harush K., Davidov G., Levin M., et al. , 2011. Self-recognition mechanism of MamA, a magnetosome-associated TPR-containing protein, promotes complex assembly. Proc. Natl. Acad. Sci. USA 108: E480–E487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y. H., Zhao Z. G., Zhou J. W., Jiang L., Bian X. F., et al. , 2011. Fine mapping of a gene responsible for pollen semi-sterility in hybrids between Oryza sativa L. and O. glaberrima Steud. Mol. Breed. 28: 323–334. [Google Scholar]
- Zhao Z. G., Jiang L., Zhang W. W., Yu C. Y., Zhu S. S., et al. , 2007. Fine mapping of S31, a gene responsible for hybrid embryo-sac abortion in rice (Oryza sativa L.). Planta 226: 1087–1096. [DOI] [PubMed] [Google Scholar]
- Zhao Z. G., Zhu S. S., Zhang Y. H., Bian X. F., Wang Y., et al. , 2011. Molecular analysis of an additional case of hybrid sterility in rice (Oryza sativa L.). Planta 233: 485–494. [DOI] [PubMed] [Google Scholar]
- Zhu S., Wang C., Zheng T., Zhao Z., Ikehashi H., et al. , 2005. A new gene located on chromosome 2 causing hybrid sterility in a remote cross of rice. Plant Breed. 124: 440–445. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The authors state that all data necessary for confirming the conclusions presented in the article are represented fully within the article.