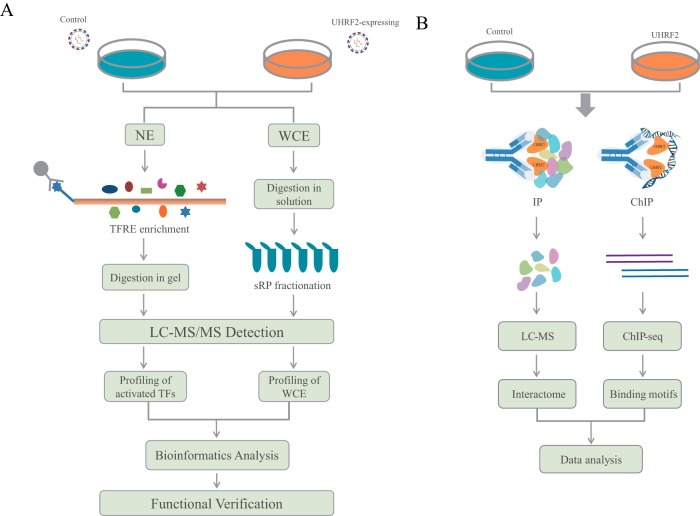
Fig. 1.
Experimental workflow for dissecting UHRF2 functions with multidimensional proteomics. A, Experimental strategy of catTFRE and profiling analysis. Gastric cancer cells were infected with mock or UHRF2 overexpression lentivirus. Control and UHRF2-OE stable cells were collected to extract nuclear extracts (NEs) and whole cell extracts (WCE) which were used for catTFRE enrichment and whole proteome profiling, respectively. MS data from different proteomics measurements were subjected to further bioinformatics analysis. Functional validation experiments were performed based on the information and clues provided by proteomics results. B, Experimental setup of UHRF2 interactome analysis and DNA binding motif analysis. Equal amount of NEs from control and UHRF2-OE cells were subjected to IP experiments using UHRF2 antibody followed by LC/MS analysis. UHRF2-OE cells were collected to do ChIP experiments with IgG and UHRF2 antibody followed by DNA sequencing.