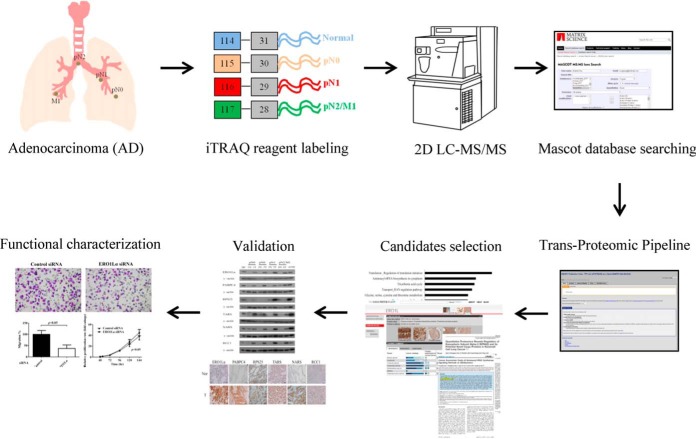
Fig. 1.
Experimental workflow used to identify potential lung ADC biomarkers from cancer tissues with different extents of LN involvement. Total proteins extracted from lung tissues with different extents of LN involvement (pN0, pN1, and pN2/M1) and from adjacent normal tissue (Nor) were digested and labeled with 4-plex iTRAQ reagents (114–117) as indicated. The labeled peptides were mixed in equal amounts and analyzed using 2D LC-MS/MS analysis. The Mascot search engine and Trans-Proteomic Pipeline were used to identify and quantify the proteins. Based on a MetacoreTM pathway analysis, the protein expression profiles released in the public Human Protein Atlas database, literature search and novelty, the differentially expressed proteins were selected and validated in clinical specimens through immunohistochemical staining and Western blot analyses. The potential biomarkers were functionally characterized using lung cancer cell lines.