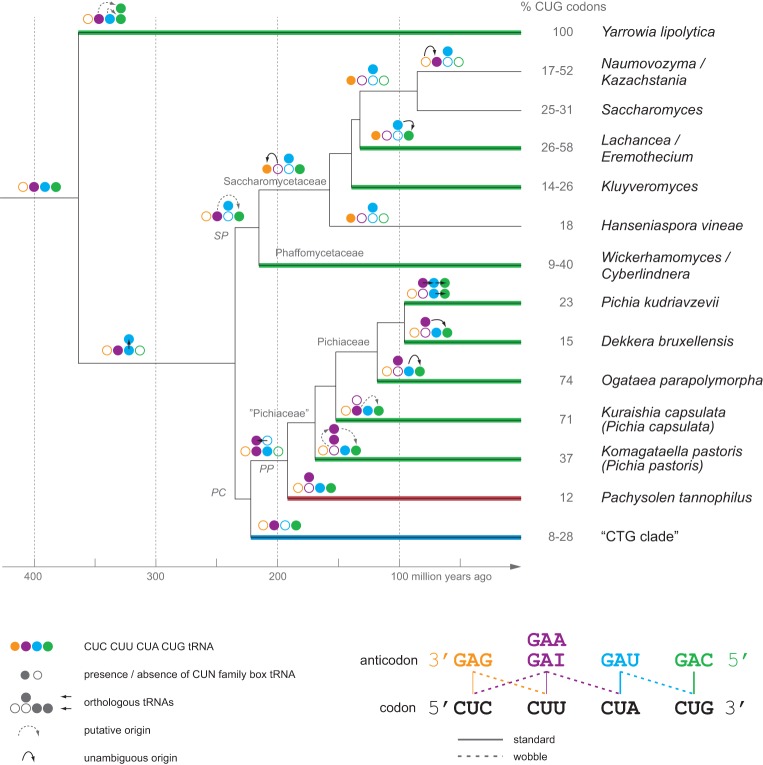
Figure 3.
The history of CUG-codon reassignment in yeasts. Decoding and origin of the tRNACAGs were plotted onto a time-calibrated yeast phylogeny adapted from Mühlhausen and Kollmar (2014). Colored lines denote the different tRNACAG types, with green representing , blue denoting the “CTG clade” , red marking the Pachysolen , and black denoting the absence of a tRNACAG in the respective branch. The schemes with the four circles represent the reconstructed CUN tRNA family at each branch, with filled and empty circles denoting the presence and absence, respectively, of the CUN family box tRNAs. Arrows indicate tRNA gene duplications followed by anticodon mutations, except for the duplication in the ancestor of the SP–PC branches that resulted in orthologous subtypes. Branches with identical decoding schemes have been collapsed. The percentage of CUG codons per species/taxon were derived from Mühlhausen and Kollmar (2014). “Pichiaceae,” as shown here, is not a commonly agreed taxon. Because all species of the respective branch in the presented tree have at least one synonymous name starting with “Pichia,” we termed the entire branch “Pichiaceae” for simplicity. The scheme at the bottom shows how the four CUN codons can be decoded by several combinations of isoacceptor tRNAs using standard and wobble base-pairing.