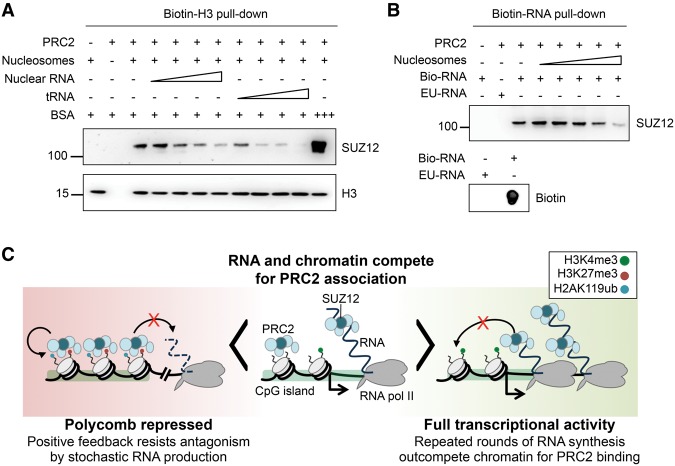
Figure 7.
RNA and chromatin compete for PRC2 binding in vitro. (A) Pull-down of PRC2 (13 ng/µL, 49 nM) by biotinylated nucleosomes (21 ng/µL, 100 nM) in the absence and presence of purified nuclear RNA or tRNA (both titrated from 0.5–65 ng/µL). BSA at 330 ng/µL (5 µM) was used to control for nonspecific occlusion effects. (B, top) Pull-down of PRC2 (13 ng/µL, 49 nM) by 2.5 µg of biotinylated nascent RNA (bio-RNA) in the absence and presence of reconstituted nucleosomes (titrated from 0.024–2.4 µM). 5-Ethynyl uridinylated RNA (EU-RNA), the precursor in the biotinylation reaction, was used as control. (Bottom) Dot blot of input bio- and EU-RNA with streptavidin-HRP. (C) Our data support a model in which PRC2 can bind either to chromatin or to RNA in a mutually antagonistic fashion. At lowly expressed genes, there is active competition between chromatin and RNA for PRC2 binding (center). At highly active genes (right), repeated rounds of nascent RNA synthesis outcompete chromatin for PRC2 binding, protecting genes from inappropriate silencing. The loss of RNA upon gene repression allows PRC2 recruitment to chromatin (left). Positive feedback mediated by EED binding to H3K27me3, and between PRC1 and PRC2, allows for stable chromatin association that resists the loss of PRC2 to RNA caused by low level or stochastic transcriptional activation. This bistable state therefore buffers changes due to transcription noise and may ensure that only robust changes in transcriptional activity are propagated as changes in epigenetic state.