Fig. 1.
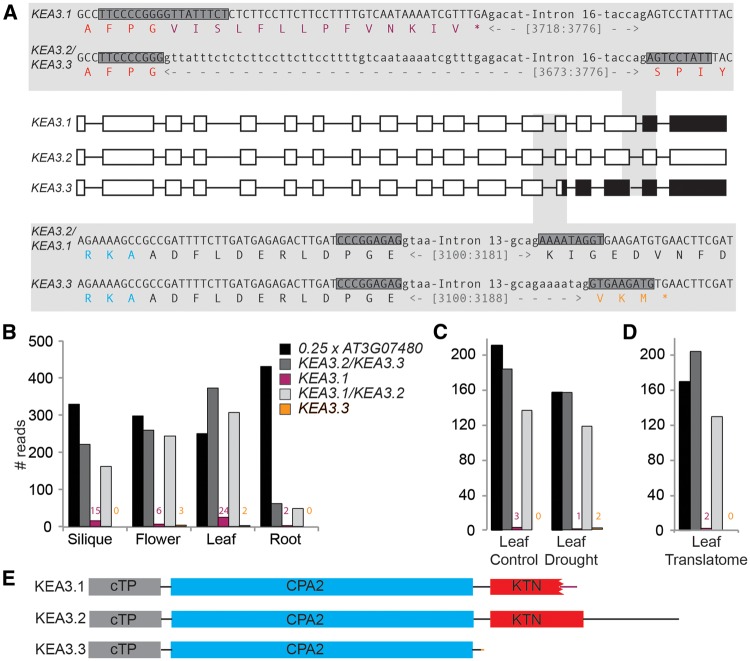
Alternative splicing events yield three KEA3 isoforms with differences in the putative C-terminal regulatory domain. (A) Overview and sequence details of the three KEA3 splice forms. Translated exons are depicted as white boxes, untranslated exons as black boxes and introns as black lines. Alternative splicing of intron #16 (upper shaded panel) yields KEA3.1 and KEA3.2/KEA3.3. Alternative splicing of intron #13 (lower shaded panel) yields KEA3.1/KEA3.2 and KEA3.3, respectively. Exons and introns are written in upper case and lower case, respectively. The encoded protein sequence is given as the single letter amino acid code. Amino acids of the KTN and CPA2 domains are colored red and blue, respectively. Amino acids specific for KEA3.1 and KEA3.3 are colored purple and orange, respectively. The 18-mers used for probing RNA-seq data for the specific splice variants are shaded dark gray. (B−D) KEA3.2 is the major splice form in all plant organs (B), under mild drought stress (C) and associated with ribosomes (D). Publicly available RNA-seq data were probed for the presence of splice form-specific 18-mers (marked by dark gray boxes in A) and a control (At3g07480). (E) Protein models of all three KEA3 isoforms.