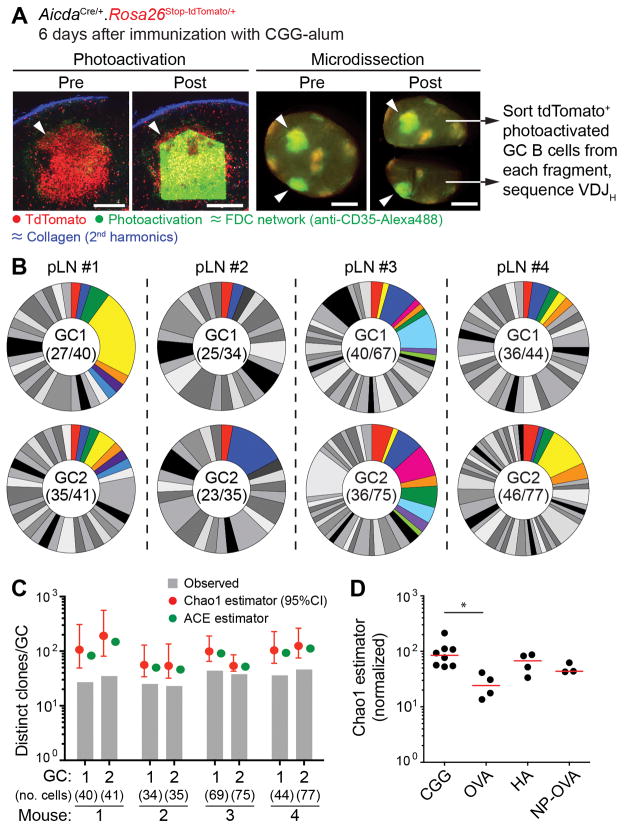
Figure 2. Clonal diversity in early GCs.
(A) Photoactivation of single early GC clusters. photo activatable-GFP-transgenic. AicdaCre. Rosa26lox-stop-lox-tdTomato mice were immunized in the footpad with 10 μg CGG in alum and imaged 6 days later. FDC networks were marked by injection of labeled antibody to CD35. Left panels show images of a single tdTomato+ cluster (arrowheads) within a pLN prior to and after photoactivation (Scale bar, 200 μm). Right panels show dissection of a single pLN with two photoactivated GCs (arrowheads) into two fragments, each of which is separately processed for cell sorting (Scale bar, 500 μm). (B) Pie charts showing clonal diversity in early GCs. Each slice represents one clone. Colored slices represent clones that were found in both GCs (upper and lower pie charts) from the same pLN. Numbers in the center of each chart are (number of clones observed/total number of cells sequenced). Clonal identity was assigned based on Igh sequence. Pairs are from 4 different mice in 3 independent experiments. (C) Estimation of total clonal richness in individual GCs using the Chao1 and ACE estimators. Graphs show observed clonal richness (from panel (B)), and total richness according to the indicated estimator. (D) Estimated clonal richness (Chao1) in GCs elicited by various antigens. Mice were immunized with 10 μg of the indicated antigen, and imaged/photoactivated as in (A). Each symbol represents one GC, bar indicates median. For comparison purposes, estimates are normalized by interpolation to the size of the smallest sample (34 cells). Note that numbers for CGG GCs in this panel differ from those in panel (C) due to normalization. Further details in Fig. S3. * p < 0.05, Kruskall-Wallis test with Dunn’s post-test. All other comparisons were not significant.