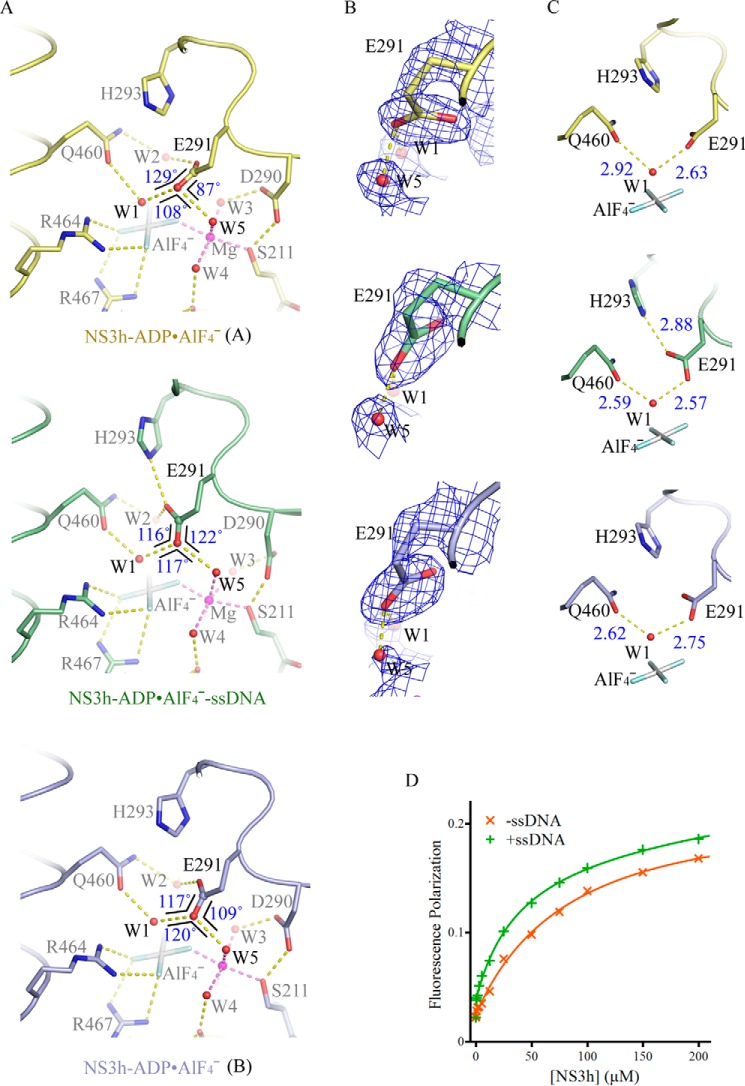
FIGURE 3.
Active-site arrangement with and without ssDNA. A, active-site arrangement of the NS3h-ADP·AlF4− complexes and NS3h-ADP·AlF4−-ssDNA ternary complex. The active-site residues and ATP mimics are modeled by sticks. The water molecules (W) and magnesium ions (Mg) are modeled by spheres. The non-carbon atoms are color-coded according to elements defined in PyMOL. The carbon atoms are colored yellow (complex A), light blue (complex B), and green (ternary complex). The yellow dashed lines represent probable atomic interactions. The three angles centered on the carbonyl oxygen are schematically highlighted by black lines, with sizes of the angles noted. The Glu-291 carbonyl groups involved in water coordination are highlighted by spheres. B, comparison of the carboxylate groups of Glu-291 in the presence and absence of ssDNA. The simulated annealing omit composite maps were contoured at 1.5 σ. C, coordination of the nucleophilic waters (W1). The structures in B and C are presented as those in A. D, binding of fluorophore-labeled GDP to NS3h. NS3h was used to titrate BODIPY-labeled GDP in the presence of NaF, AlCl3, and MnCl2. To analyze the impact of nucleic acid enhanced transition-state mimic binding, ssDNA (dT12) was added. Fluorescence polarization values derived from single measurements were plotted against NS3h concentrations.