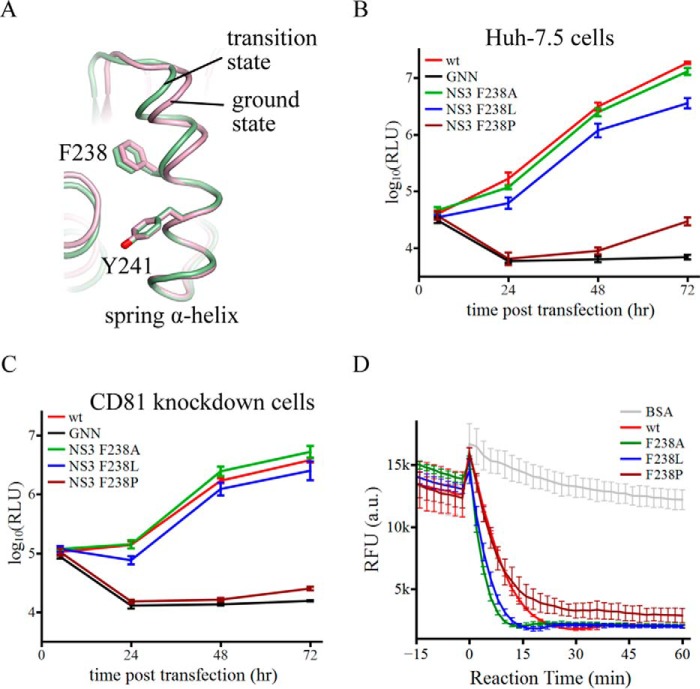
FIGURE 5.
The α-helical structure of the spring helix is important for virus replication. A, schematic presentation of the spring helices in the ground-state (pink) and transition-state ternary complexes (green). The structures were aligned through amino acids 240–245. Phe-238 targeted by mutagenesis and Tyr-241 are modeled by sticks. B, replication of reporter HCV viruses in Huh-7.5 cells. The wild-type (wt) virus is Jc1(p7-nsGluc2A). The GNN strain is replication incompetent due to asparagine substitution of two catalytic aspartate residues in motif VI of NS5B (the RNA dependent RNA polymerase). F238A, F238L, and F238P were introduced in the NS3 helicase domain. Virus replication was measured by accumulation of secreted luciferase activities. Every data point was averaged from three independent assays. The error bars represent S.D. C, replication of the same set of reporter HCV viruses in Huh-7.5 CD81 knockdown cells. The data are plotted as those in B. D, duplex unwinding activities of the wild-type and mutant NS3 helicase proteins. The unwinding reactions were monitored by fluorescence of a molecular beacon annealed in a duplex. Separation of the duplexes led to self-annealing of the molecular beacons and, therefore, a decrease of fluorescence. Fluorescence intensities were measured every 2 min at 37 °C. The presented data sets, which were averaged form triplicate assays, cover 15 min without ATP and 60 min with ATP. The error bars are S.D. The negative control used bovine serum albumin (BSA).