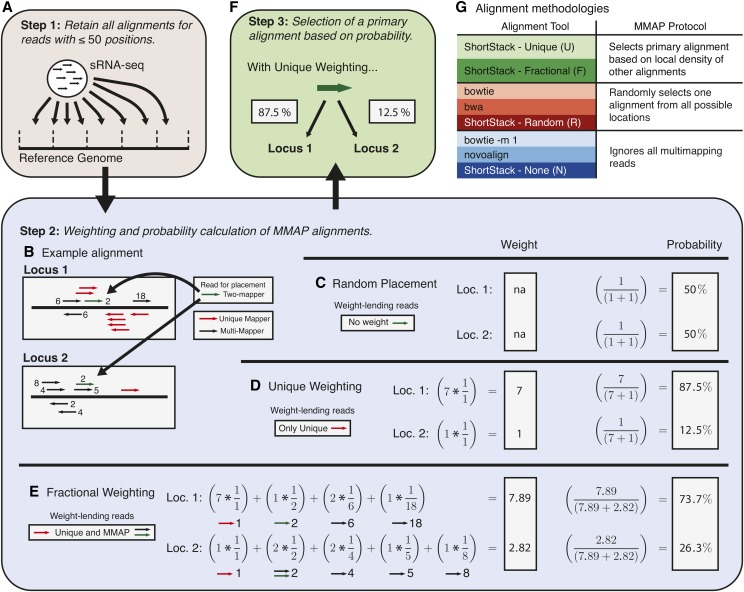
Figure 2.
ShortStack3 alignment methodology. (A) First step in alignment by ShortStack: initial alignment of sRNA-seq reads to a reference genome. (B) Example of local alignments for a read (green) with an MMAP-value of two. Numbers adjacent to reads indicate their MMAP-value. (C) Weighting scheme for random placement of MMAP reads. (D) Weighting scheme for ShortStack’s Unique (U) method. (E) Weighting scheme for ShortStack’s fractional (F) method. (F) Final step: choosing a single primary alignment based on calculated probabilities. (G) Alignment tools grouped by MMAP methods.