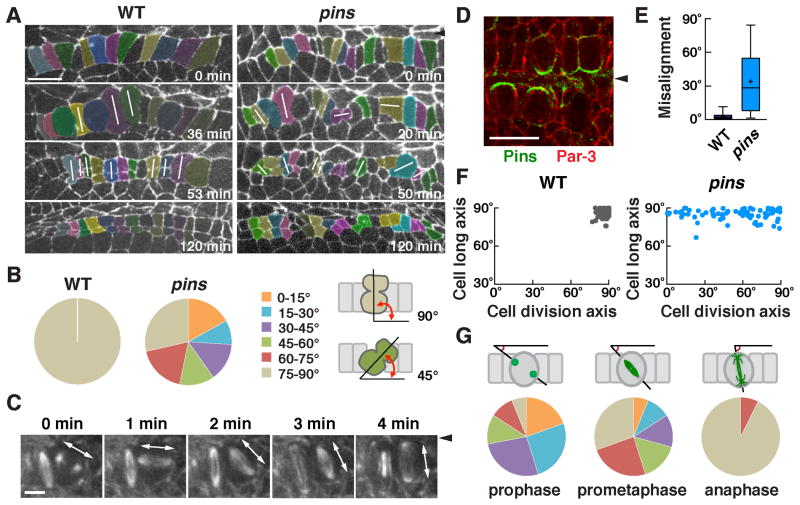
Figure 3. Pins/LGN is asymmetrically localized and is required to orient cell division during square grid formation.
(A) Stills from time-lapse movies of wild-type and pins mutant embryos expressing β-catenin:GFP. White lines indicate cells in anaphase or telophase. (B) Quantification of the cell division axis (0° is parallel to the ventral midline). All wild-type cells divided at 75–90° relative to the midline. Cell divisions were frequently misoriented in pins mutants (p < 0.001) (Chi square test). (C) Stills from a time-lapse movie of wild-type cells expressing Jupiter:mCherry. One spindle rotates 90° in the epithelial plane. (D) Pins/LGN accumulates at cell interfaces contacting midline cells in Phase II (stage 12). (E) The gap between the cell division axis and the cell long axis was significantly larger in pins mutants (p < 0.001) (unpaired t test). Boxes, 25th to 75th percentile; whiskers, 2.5th to 97.5th percentile; horizontal line, median; +, mean. Plot shows the distribution of average values across embryos. (F) The cell division axis and the cell long axis were both close to perpendicular to the ventral midline in wild type. The orientation of the cell long axis in pins mutants (85°±0.5°) was similar to wild type (86°±0.4°), but cell division occurred at a much wider range of orientations. (G) Angular distributions of the aligned centrosomes at prophase and spindles at prometaphase or anaphase. n = 73–77 cells in 3 embryos/genotype in B, E, and F and 82 cells in 5 embryos in G. Arrowheads, ventral midline. Bars, 10 μm in A,D, 5 μm in C. See also Figure S2.