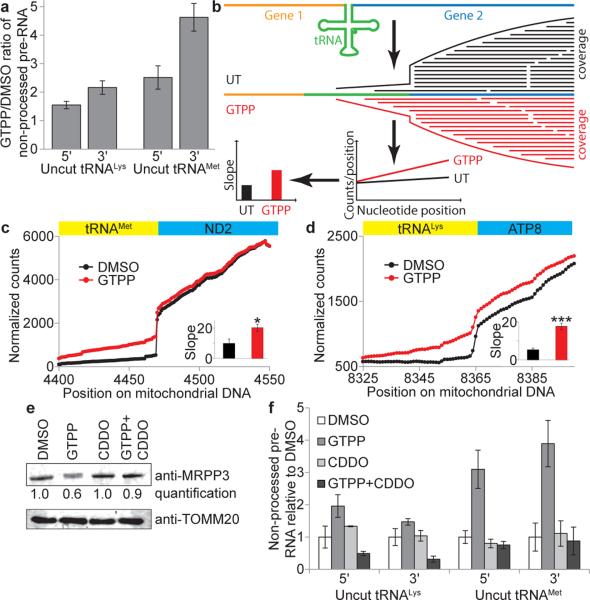
Fig. 3. Mitochondrial pre-RNA processing defects upon UPRmt.
a, qPCR of mitochondrial pre-RNA at tRNAMet and tRNALys RNAseP processing sites upon induction of UPRmt with GTPP (6h). Error bars, averages ±s.d. (n=3 biological replicates). b, RNA-seq for analysis of mitochondrial pre-RNA processing defects based on number of reads crossing the tRNA/mRNA gene junction. Slope of coverage in the tRNA gene adjacent to the cut site is used as a measure of processing. c-d, Normalized RNA-seq coverage across tRNA/mRNA gene borders for tRNAMet (c) and tRNALys (d) with average of slopes (±s.d.) from b indicated in the inset (n=3 biological replicates, two-tailed p-values *p≤0.05, ***p≤0.001).e, Quantitative western blot analysis of MRPP3 levels upon treatment of cells with DMSO, GTPP, CDDO, or GTPP + CDDO co-treatment for 6 hours. f, Mitochondrial pre-RNA accumulation upon co-treatment with GTPP and CDDO (as in a). Data are average values ±s.d. (n=3 biological replicates). For gel source data, see Supplementary Figure 1.