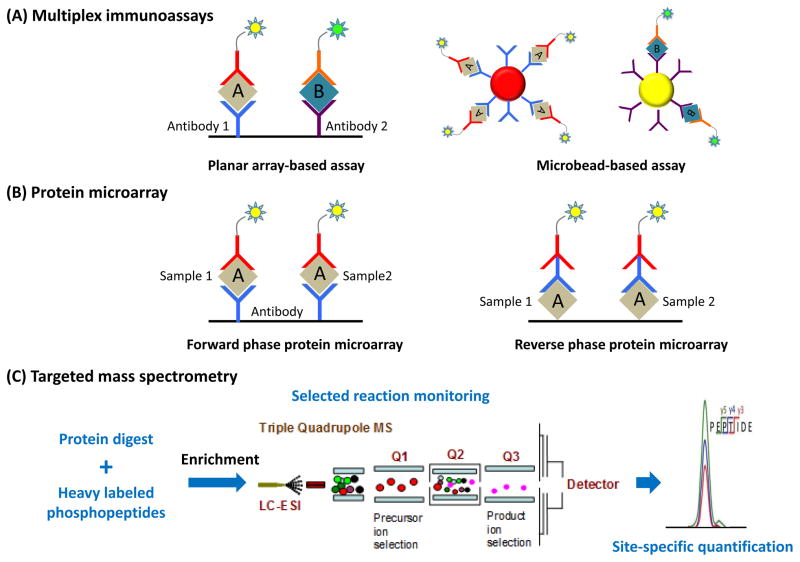
Figure 2. Overview of targeted approaches for phosphoproteomics.
(A) Multiplex immunoassays. In planar array-based assay, or ELISA, Specific primary phospho-antibodies are anchored to a carbon electrode plate (Antibody-1, and -2), which recognize phosphoproteins of interest. With the application of a fluorescent-tagged secondary antibody, the fluorescence signal measured corresponds to the expression level of that particular phosphoprotein in the sample. In the microbead-based array, primary antibodies are conjugated to the microspheres, and each microsphere batch is distinctly barcoded with different fluorophores mixed at a specific ratio (red, and yellow beads). (B) Protein microarray (or protein chip).In the forward-phase protein microarrays (FPPAs), phospho-antibodies are anchored on the chip to detect the phosphoproteins of interest; whereas in the reverse-phase protein microarrays (RPPAs), the clinical samples are directly spotted on the surface. With the sequential incubation of the cognate primary and secondary antibodies, phosphoprotein of interest could then be detected. (C) Targeted mass spectrometry. Heavy isotope-labeled synthetic peptides are typically spiked into protein digest as internal standards prior to IMAC/MOAC enrichment. The enriched phosphopeptides are subjected to SRM/PRM based targeted quantification, and the levels of phosphorylation are quantified based on light-to-heavy isotopic ratios.