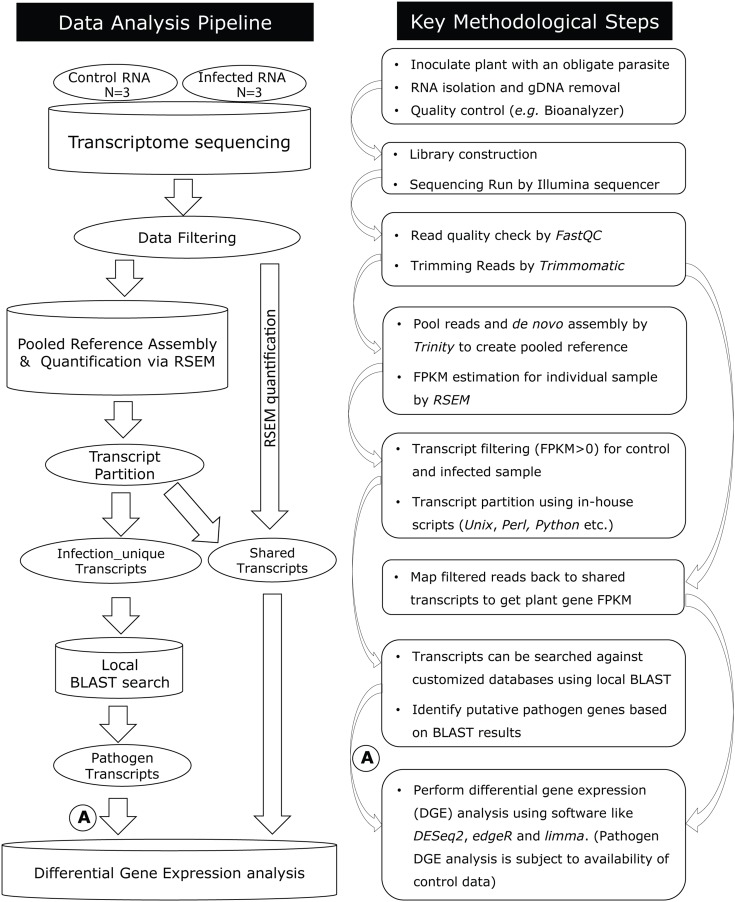
FIGURE 1.
Diagram summarizing the data analysis pipeline to analyze host–pathogen metatranscriptomes and key methodological steps. A step-by-step protocol of this pipeline is available (Supplementary Material). After quality filtering, RNA-seq reads are assembled de novo using Trinity. For pathogen transcript discovery, a “pooled reference” is assembled combining control and infected plant reads, which are further divided into control-unique, infected-unique, and shared groups. For plant differential gene expression analysis, shared transcripts are used as a reference, against which control and infected reads are mapped by RSEM. A = DGE analysis for pathogen transcripts are subject to availability of a reference sample.