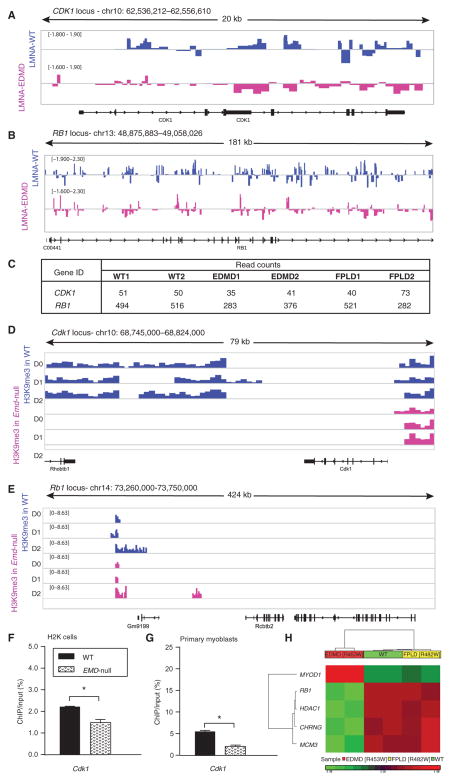
Fig. 2. Cell cycle loci show decreased association with EDMD lamin A and inadequate heterochromatin formation in differentiating EDMD myogenic cell.
Lamina association of cell cycle loci assessed by DamID-seq showed reduction in binding of mutated lamin A, which resulted in reduced heterochromatinization of these loci as assessed by ChIP-seq in emerin-null murine myogenic cells (H2K cells). (A and B) Wiggle track format (Wig) of lamin A enrichment on two cell cycle genes, (A) CDK1 and (B) RB1. Data are presented as log2 of DamLMNA-associated genomic regions/DamOnly genomic regions. (C) Normalized read counts of lamin A peaks at the CDK1 and RB1 loci. (D and E) Wig tracks of H3K9me3 enrichment at Cdk1 (D) and Rb1 (E) loci. (F and G) ChIP-qPCR validation of H3K9me3 enrichment at the Cdk1 locus in WT emerin and emerin-null (EMD-null) (F) murine H2K cells and (G) primary myoblasts from extensor digitorum longus (EDL) murine muscle. Data are means ± SEM. (H) DNA methylation analysis of human skeletal muscle cells infected with lentivirus encoding the WT LMNA gene or LMNA genes with one of two different mutations [WT, EDMD (p.R453W), and FPLD (p.R482W)] focused on genes previously implicated in EDMD pathology (3). Heat map of genes that show statistically significant differences (±1.2 FC; P < 0.05, FDR-corrected) between WT and EDMD and WT and FPLD cells. chr, chromosome.