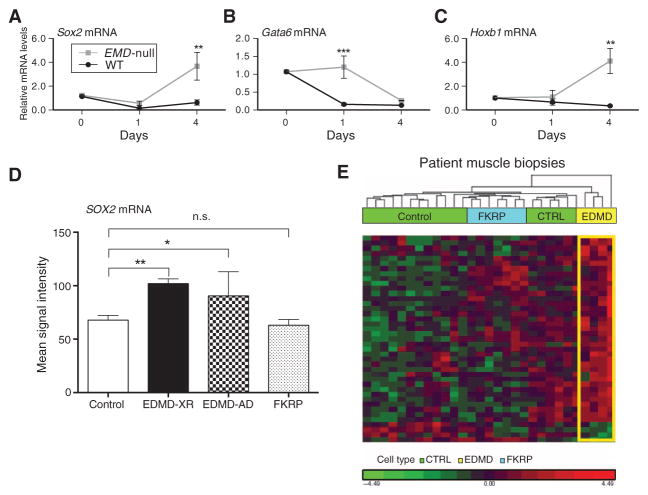
Fig. 5. Sox2 and downstream target genes show abnormal gain of function in differentiating emerin-null myogenic cells and in muscle biopsies from EDMD patients.
(A to C) Differentiating WT and emerin-null H2K cells were studied at d0, d1, and d4 after induction of myogenic differentiation. qRT-PCR of (A) Sox2 and two downstream transcriptional target genes [Gata6 (B) and Hoxb1 (C)] showed down-regulation in WT cells, whereas emerin-null cells show abnormally high expression of all three genes. Two-way ANOVA with repeated measures, followed by Bonferroni’s multiple comparisons test, ***P < 0.0005 and **P < 0.005; n = 3. Error bars indicate ± SEM. (D and E) mRNA profiles were accessed (3) by Affymetrix microarrays in muscle biopsies taken from control, EDMD, and FKRP patients [control (CTRL), n = 13; EDMD-XR, n = 4; EDMD-AD, n = 3; FKRP, n = 7]. (D) Affymetrix signal intensity query of Sox2 mRNA showed abnormal up-regulation in both EDMD-XR and EDMD-AD patient muscle relative to control samples. (E) Hierarchical clustering of mRNA transcripts corresponding to H3K9me3 ChIP-seq loci that displayed specific decreases, relative to WT, in heterochromatin in emerin-null H2K myogenic cells (n = 42). The large majority of these murine ChIP-seq loci showed up-regulation of the corresponding mRNA transcripts in EDMD patient muscle biopsies (yellow) relative to control (green) and disease control FKRP (blue). n.s., not significant.