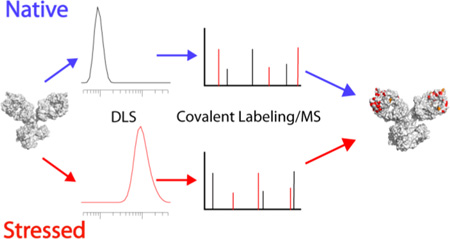
Abstract
Protein therapeutics are rapidly transforming the pharmaceutical industry. Unlike for small molecule therapeutics, current technologies are challenged to provide the rapid, high resolution analyses of protein higher order structures needed to ensure drug efficacy and safety. Consequently, significant attention has turned to developing new methods that can quickly, accurately, and reproducibly characterize the three-dimensional structure of protein therapeutics. In this work, we describe a method that uses diethylpyrocarbonate (DEPC) labeling and mass spectrometry to detect three-dimensional structural changes in therapeutic proteins that have been exposed to degrading conditions. Using β2-microglobulin, immunoglobulin G1, and human growth hormone as model systems, we demonstrate that DEPC labeling can identify both specific protein regions that mediate aggregation and those regions that undergo more subtle structural changes upon mishandling of these proteins. Importantly, DEPC labeling is able to provide information for up to 30% of the surface residues in a given protein, thereby providing excellent structural resolution. Given the simplicity of the DEPC labeling chemistry and the relatively straightforward mass spectral analysis of DEPC-labeled proteins, we expect this method should be amenable to a wide range of protein therapeutics and their different formulations.
Graphical Abstract
Protein therapeutics are the fastest growing segment of the pharmaceutical market, accounting for one-third of the overall late-stage drug development pipeline. They are anticipated to represent 20% of the total pharmaceuticals market value by 2017.1 One key element in ensuring the safety and efficacy of these biologic drugs is the ability to measure and control the three dimensional (3D) structure of the protein active ingredients. In contrast to more traditional small molecule therapeutics, however, obtaining accurate, high resolution measurements of protein structures has proven to be extremely challenging.
Current structural techniques fall into two major categories: (1) rapid, low resolution techniques and (2) time and sample intensive, high resolution techniques.2 Intrinsic fluorescence, circular dichroism (CD), dynamic light scattering (DLS), differential scanning calorimetry (DSC), and activity assays are examples of the first type. These methods provide an ensemble average of structures or are sometimes insensitive to certain structural changes. NMR and X-ray crystallography are important examples of powerful high resolution techniques, but these methods are time-consuming, require a large amount of protein, and are not amenable to all proteins. Thus, there is a growing need for other techniques that can provide better resolution than the first category of techniques but do so in way that is easier and faster than the second category of techniques. This need is especially pressing as the field of protein therapeutics expands, and as the ability to ensure that the 3D structures of proposed biosimilars are the same as the original branded drug becomes a major issue.3–5
Mass spectrometry (MS)-based techniques offer an alternative because they can be rapid, provide moderate resolution, and can be sample efficient. Accordingly, these techniques have begun to fill an important niche in protein therapeutic analyses. The primary techniques used for monitoring protein solution structure by MS are hydrogen/deuterium exchange (HDX), chemical cross-linking, and covalent labeling. In HDX the mass spectrometer is used to measure the exchange of amide hydrogens for deuterium (or vice versa), and the extent of exchange at individual sites provides an indication of solvent accessibility and protein dynamics near that site. HDX/MS has been widely used to analyze protein structure6–10 and recently has been applied to characterize the structure of protein therapeutics.11–14 One challenge associated with HDX/MS is the transient nature of the label. As a result, special care and often expensive instrumentation are required to minimize back exchange and to accurately locate deuterated sites.
Methods that use covalent bond formation to characterize protein structure are not subject to back exchange. They also provide complementary information by reporting on protein side chains. Chemical cross-linking typically uses bifunctional reagents to link residues that are spatially adjacent despite being distant in linear sequence. The cross-linked peptides are then sequenced and identified by MS, thereby revealing nearby residues. This method has been used to probe the structures of individual proteins15 and protein complexes.16–19 While this technique is not commonly used to study protein therapeutics, it has been used for antibody epitope mapping.20 Other covalent labeling techniques use monofunctional reagents to monitor residue solvent accessibility as a means of probing structure. Hydroxyl radical footprinting (HRF) is the most common of these techniques.21–24 In this method, hydroxyl radicals are produced through radiolysis or photolysis of water or hydrogen peroxide, and the resulting radicals then oxidize solvent accessible sites on the protein. Because of its broad reactivity and success with other protein systems, HRF has recently been applied to monitor structural changes in therapeutic proteins.25,26 The technique was shown to be quite sensitive to subtle structural changes as it was able to distinguish expired protein therapeutics from fresh ones.25 HRF also demonstrated the ability to identify the regions of aggregation in therapeutic monoclonal antibodies (mAbs).26 While HRF shows great promise for studying therapeutic proteins, there are some challenges associated with implementation. Most notably, oxidation by hydroxyl radicals can produce over 50 different types of modifications, which can complicate MS analysis.23 Moreover, in its most commonly used forms, a laser or synchrotron source is necessary to generate the radicals, which adds complexity and limits its wide applicability.
Another approach to covalent labeling uses amino acid-specific reagent molecules to modify solvent exposed residues. A wide range of reagents are available, ranging from those that have narrow specificity (e.g. succinimides) to those that have broad reactivity (e.g. DEPC).27 This approach to labeling is simple as it requires no specialized equipment and typically produces only a single type of product, facilitating mass spectral analysis. While amino acid-specific reagents have been widely used to probe monomeric and oligomeric proteins,27 their application to therapeutic proteins has been very limited. To our knowledge, only the carboxylate-specific reagent pair of 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC) and glycine ethyl ester (GEE) has been used to probe the structure of a mAb.28 This particular labeling chemistry is relatively simple to implement; however, because it is limited to only Asp and Glu residues, it results in relatively poor coverage of the protein’s surface area and thus low effective structural resolution. Another reagent with broader reactivity such as DEPC should maintain the simplicity of this type of covalent labeling, while at the same time increasing resolution. Because DEPC is capable of labeling all nucleophilic residues, our group has shown that this reagent is capable of monitoring approximately 30% of surface residues of the average protein.29,30 Such broad reactivity has enabled this reagent to provide insight into protein structure as well as protein-metal and protein-protein interactions.27,30–34
In this work, we demonstrate the ability of DEPC labeling to assess structural perturbations in protein therapeutics by investigating three proteins before and after stressed conditions. We find that covalent labeling is capable of identifying stress-induced structural perturbations in protein therapeutics, including the interface through which the protein therapeutics aggregate.
EXPERIMENTAL METHODS
Materials
Diethylpyrocarbonate (DEPC), imidazole, iodoacetamide, L-cysteine, papain from papaya latex, tris(2-carboxyethyl)phosphine (TCEP), and DL-dithiothreitol (DTT) were obtained from Sigma Aldrich (St. Louis, MO). The mAb immunoglobulin G1 (IgG1) was purchased from Waters Corporation (Milford, MA). Human β-2-microglobulin (β2m) was obtained from Fitzgerald Industries International (Concord, MA). Recombinant Human Growth Hormone (HGH) was purchased from Biovision (San Francisco, CA). Urea was purchased from Acros Organics (Geel, Belgium). Both immobilized trypsin and chymotrypsin were obtained from Princeton Separations (Adelphia, NJ). Sodium phosphate monobasic monohydrate was purchased from EM Science (Darmstadt, Germany). Sodium phosphate dibasic anhydrous, hydrogen peroxide, methanol, formic acid, acetonitrile, and water were purchased from Fisher Scientific (Fair Lawn, NJ). Centricon molecular weight cutoff (MWCO) filters were obtained from Millipore (Burlington, MA).
Sample Preparation
β2m and IgG1 were prepared in 50 mM ammonium acetate (pH 7.4) and 50 mM sodium phosphate buffer (pH 7.4), respectively. Both proteins were then incubated at 75 °C for 15 min (IgG) or 1 day (β2m) for thermal degradation conditions. Oxidative conditions were carried out by incubating the protein in the presence of 3% H2O2 (w/w) at room temperature for 1 day. HGH was prepared in 10 mM sodium phosphate buffer (pH 8.0), and incubated at 65 °C for 2, 12, and 24 hours. After the forced degradation conditions, the proteins were reacted with DEPC and then analyzed by MS.
DEPC Labeling Reactions
Stock solutions of DEPC were prepared in acetonitrile. The DEPC reactions of β2m were performed for 1 min at 37 °C and were initiated by adding DEPC in a molar excess of 2.5. The total reaction volume for the experiments was 100 µL, and the total amount of acetonitrile added was 1%. Based on our previous work, this low percentage of acetonitrile has no noticeable influence on protein structure.30–33 Experiments with β2m were performed in triplicate. The reactions were quenched after 1 min by adding 10 mM imidazole. Labeling of IgG1 (5 µM) was performed at a protein to DEPC molar ratio of 1:4 in a 50 mM phosphate buffer at pH 7.4. The solution was reacted for 5 min at room temperature before quenching by the addition of imidazole at a 1:50 (DEPC:imidazole) ratio. Five replicate reactions and analyses were conducted on the IgG1 samples. DEPC labeling of HGH was performed at a 1:5 (protein:DEPC) ratio for 1 min at room temperature. The amount of acetonitrile added was 1%. The reaction was quenched by the addition of imidazole at an 80 molar excess to DEPC. Three replicate reactions and analyses were conducted on the HGH samples.
Proteolytic Digestion
Digestion was performed with immobilized chymotrypsin for B2m and immobilized trypsin for HGH. To achieve complete digestion of IgG1, an initial digestion with activated papain was necessary. This was then followed by digestion with immobilized trypsin. For more details about the digestion conditions, please refer to the supplemental information.
Mass Spectrometry
On-line high performance liquid chromatography (HPLC) MS analyses were performed on all protein digests. The HPLC details can be found in the SI. Mass analysis of β2m proteolytic fragments was carried out on a Bruker AmaZon (Billerica, MA, USA) quadrupole ion trap mass spectrometer equipped with an electrospray ionization source. Typically, the electrospray needle voltage was kept at ~4 kV, and the capillary temperature was set to 250 °C. Either collision-induced dissociation (CID) or electron transfer dissociation (ETD) was used to obtain tandem mass spectra.
For IgG, MS analyses were performed using a Thermo Scientific Orbitrap Fusion (Tewksbury, MA) mass spectrometer. The electrospray ionization source was typically operated at a needle voltage of 2200 volts, and the ion transfer tube temp was set to 300 °C. Tandem mass spectra were collected using CID with a normalized collision energy of 35%. Due to the large number of measured peaks, an exclusion limit of 60 sec was activated after five spectra were collected for any given peak. The resolution of the Orbitrap was set to 60000.
MS analysis for HGH was performed using a Thermo Scientific LTQ-XL Orbitrap (Tewksbury, MA) mass spectrometer equipped with an electrospray ionization source. The ESI needle voltage was kept at 5 kV. Tandem mass spectra were generated using an HCD collision energy of 35.
Peptide identification
Raw mass spectral data files were converted to .mgf format using msconvert software.35 The .mgf files were analyzed with SearchGUI.36 The search engines X!tandem,37,38 MS Amanda,39 MS-GF+,40 OMSSA,41 and Comet42 were all used. Spectra were searched against a database constructed from the cRAP database (http://www.thegpm.org/crap/index.html) with the sequence of the proteins of interest added. Spectra were searched against the custom database and against the reverse, decoy database. Variable modification by DEPC of the residues H, Y, K, T, S and the protein N-terminus was added as a user modification (mass addition of 72.0211). Variable oxidation of M was also used in searches. Carboxyamidomethylation of cysteine was used as a fixed modification. Unspecific enzyme cleavage was selected, and a precursor mass tolerance of 10 ppm was used.
Search data were visualized using Peptideshaker43 with protein, peptide and PSM FDRs set at 1%. PTMs were scored using the PhosphoRS algorithm. Identification features were exported in .csv format, and these features were used to construct a custom database for peak identification in MZmine.
Peptide peak quantification
Raw data files were imported into MZmine,44 and mass detection was done in centroid mode at the MS1 level. Chromatograms were constructed, deconvoluted and peak identification was performed with a custom database constructed from the Peptideshaker export. When multiple files were analyzed, deconvoluted spectra were aligned using the RANSAC algorithm. The quantified, identified and aligned data were exported to a .csv file using the export function. A representative workflow can be found in Figure S1 in the SI.
Biophysical Characterization of Proteins
Circular dichroism spectroscopy, fluorescence spectroscopy, dynamic light scattering, and size-exclusion chromatography were also used to characterize the proteins after heat or oxidative stress. The details of these methods can be found in the SI.
RESULTS AND DISCUSSION
β-2 microglobulin (β2m)
β2m shares the same β-sandwich fold as each of the domains in IgG and thus was chosen as an initial model system. The protein under native and both heat- and oxidatively-degraded conditions was probed using DEPC. Labeling was done at a 2.5:1 molar ratio (DEPC:protein) as previous work from our group had demonstrated that a labeling ratio of 4:1 or less provided good labeling yield without significantly perturbing a protein’s structure.30
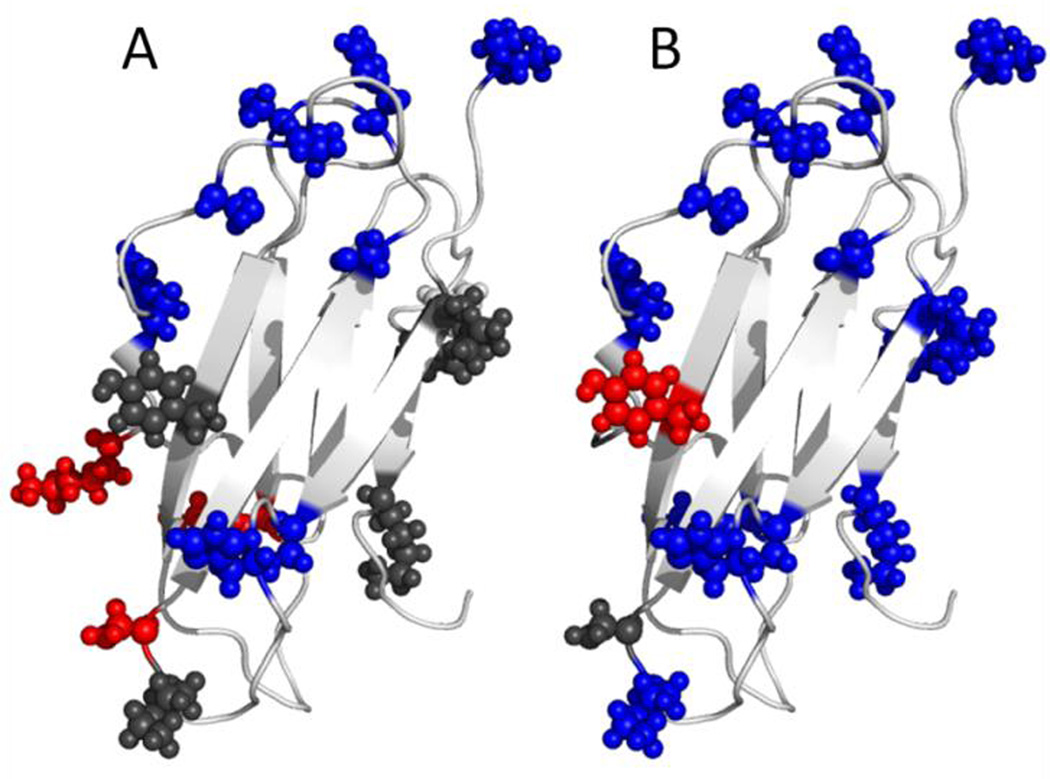
The modification results for the β2m residues under all three conditions reveal that β2m undergoes noticeable structural changes upon exposure to elevated temperature and H2O2 (Table 1). The residues that are labeled under native conditions are displayed as spheres in Figure 1A and B. All 16 of these residues are found on the exterior of the protein and are exposed to solvent, which is consistent with previous DEPC labeling results for this protein under native conditions. After being exposed to thermal stress for 24 hours, the extent and pattern of labeling significantly changes. The residues highlighted in blue in Figure 1A undergo statistically significant decreases (p-value < 0.05) in labeling extent, while the residues in red undergo an increase in labeling extent. Because many residues undergo a decrease in labeling extent and these residues are clustered on one face of the protein, these results suggest that the protein aggregates upon overnight heating. Support for this conclusion is found from SEC measurements (Figure S2 in the Supporting Information (SI)), which reveal protein aggregates are formed. The residues that decrease in labeling extent (i.e. Ile1, Ser28, His31, Ser33, Ser55, and Ser57) are likely at the interface(s) of these oligomers. Moreover, the same region of the protein is buried in the MHC complex that β2m forms physiologically.46 This region’s exposure is likely thermodynamically unfavorable in the monomeric protein, potentially explaining its propensity for aggregation at this site. Interestingly, two residues (i.e. Lys41 and Lys48) on the loop that connects two of the aggregating β strands show an increase in labeling after heating, suggesting that this region of the protein unfolds upon heating to facilitate aggregation.
Table 1.
Modification percentages for individual residues of β2m before and after heating at 75 °C for 24 hours or oxidation with 3% H2O2 for 24 hours.
| β2m | Native (control) |
Heated for 1 day |
Significant* | 3% H2O2 | Significant* |
|---|---|---|---|---|---|
| N-term | 49 ± 1 | 44 ± 2 | yes | 7.6 ± 0.3 | yes |
| Lys6 | 2.5 ± 0.1 | 3 ± 0.5 | no | 0 ± 0 | yes |
| Ser11 | 20 ± 3 | 11.8 ± 0.3 | yes | 6 ± 2 | yes |
| His13 | 32 ± 2 | 17 ± 1 | yes | 4 ± 1 | yes |
| Lys19 | 2.7 ± 0.4 | 2 ± 2 | no | 0 ± 0 | yes |
| Ser20 | 1.1 ± 0.3 | 4.8 ± 0.7 | yes | 1.7 ± 0.3 | no |
| Ser28 | 0.25 ± 0.05 | 0 ± 0 | yes | 0.3 ± 0.09 | yes |
| His31 | 1.9 ± 0.2 | 0.72 ± 0.08 | yes | 0 ± 0.1 | yes |
| Ser33 | 0.68 ± 0.09 | 0 ± 0 | yes | 0.18 ± 0.01 | yes |
| Lys41 | 0.6 ± 0.1 | 1 ± 0.1 | yes | 0 ± 0 | yes |
| Lys48 | 0 ± 0 | 1.2 ± 0.2 | yes | 0 ± 0 | no |
| His51 | 2.2 ± 0.3 | 0 ± 0 | yes | 0.27 ± 0.07 | yes |
| Ser55 | 1.4 ± 0.2 | 0 ± 0 | yes | 0 ± 0 | yes |
| Ser57/ Lys58 |
2.5 ± 0.4 | 0 ± 0 | yes | 0 ± 0 | yes |
| Tyr67/ Thr68 |
2.2 ± 0.3 | 2 ± 1 | no | 6 ± 2 | yes |
| Lys91 | 2.1 ± 0.4 | 2 ± 0.2 | no | 0.1 ± 0.2 | yes |
| Lys94 | 3.1 ± 0.6 | 3 ± 0.4 | no | 0.2 ± 0.3 | yes |
A difference was considered significant if the p-value, calculated by performing an unpaired T-test, was less than 0.05 (corresponding to a 95% confidence level, n=3)).
Figure 1.
Covalent labeling results for β2m. Spheres represent residues that were labeled with DEPC. The color indicates whether the residue has undergone any significant change in labeling after being exposed to a perturbing condition (blue: decrease, red: increase, gray: no change). A) Heating at 75°C for 24 hours. B) Oxidation with 3% H2O2 for 24 hours. Changes in covalent labeling are mapped onto the NMR structure of β2m (PDB accession code: 2XKS).
Covalent labeling of the oxidized protein also indicates that the protein aggregates after exposure to H2O2. Nearly all residues undergo a decrease in labeling extent (Figure 1B), signifying extensive aggregation. Analysis via SEC corroborates this conclusion (Figure S2 in the SI). While many of the same residues decrease in labeling as in the heated sample, the additional residues indicate the aggregates are also mediated by other interfaces. Overall, the covalent labeling experiments with β2m successfully demonstrate the ability of this technique to identify structural changes, especially interfacial sites that are formed upon heat- and oxidatively-induced aggregation.
Immunoglobulin G (IgG)
The promising results with β2m prompted us to apply the method to IgG under native and thermally degraded conditions. To minimize structural perturbations to IgG during the labeling reaction, we limited the DEPC:protein ratio to 4:1. We also monitored the protein’s structure using CD and fluorescence spectroscopy. Both techniques demonstrate that the protein undergoes no significant structural perturbations after reacting with DEPC at these concentrations (Figures S3 and S4 in the SI), confirming the structural integrity of the protein. We were pleased to find that DEPC labeling of IgG under these conditions results in the labeling of almost 30% of the amino acids in the protein (Table S1 and S2 in the SI).
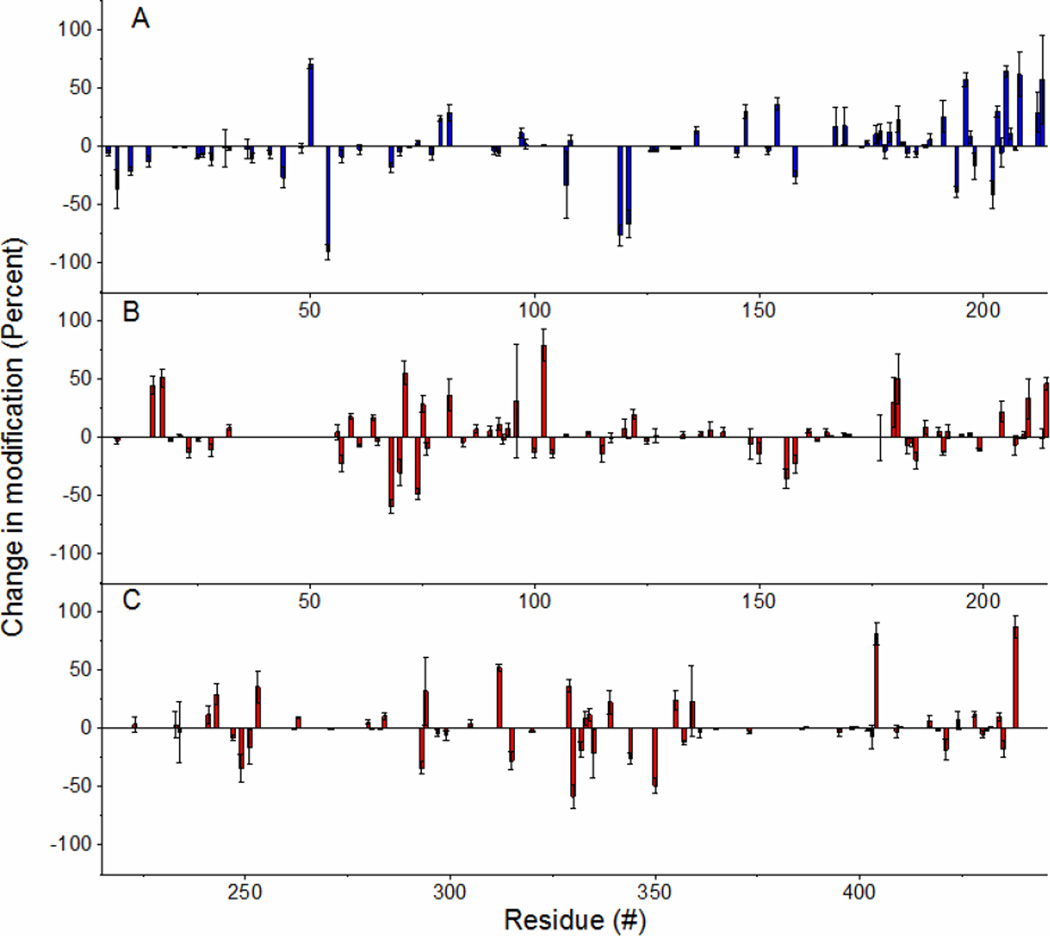
Upon comparing the labeling results of the thermally degraded sample with those generated under native conditions, we find that numerous sites undergo changes in labeling. Figure 2 summarizes these results by showing the percent change in DEPC labeling that each residue undergoes upon heat treatment relative to the unheated sample. Figure 2A illustrates the labeling for IgG’s light chain, while Figure 2B shows the results for the heavy chain. Because about 200 residues are labeled in the protein, these results were further simplified by considering only the statistically significant changes in each domain. The significant relative changes were broken into bins based on the magnitude of the change (Table 2). Each domain has a relatively equal number of residues undergoing increases or decreases in labeling; however, almost all of the residues in the light chain’s variable domain (VL) undergo significant decreases in labeling. This clustering of residues suggests that the protein might be aggregating upon heating, and this domain mediates this aggregation. Indeed, DLS demonstrates that IgG undergoes significant aggregation after 15 min of heating at 75 °C (Figure S5 in the SI).
Figure 2.
Bars represent changes in modification of IgG1 after heating from experiments involving five replicates. Negative values represent residues that are more protected after heating. A) Light chain. B) Top: VH and CH1 domain of heavy chain. Bottom: CH2 and CH3 domain of heavy chain
Table 2.
Number of residues within each domain of IgG1 whose relative labeling change after heating is statistically significant and whose value falls within the labeled bin.
| Domaina | < −80% | < −40% | < −10% | Total De- crease |
> 200% |
> 100% |
> 10% | Total In- crease |
|---|---|---|---|---|---|---|---|---|
| VL | 12 | 8 | 1 | 21 | 4 | 0 | 0 | 4 |
| CL | 10 | 3 | 3 | 16 | 12 | 3 | 2 | 17 |
| VH | 13 | 2 | 1 | 16 | 13 | 1 | 1 | 15 |
| CH1 | 9 | 2 | 1 | 12 | 13 | 4 | 2 | 19 |
| CH2 | 9 | 5 | 0 | 14 | 4 | 3 | 3 | 10 |
| CH3 | 5 | 6 | 0 | 11 | 4 | 1 | 3 | 8 |
CL and VL represent the constant and variable domains of the light chain. VH and CH1–3 represent the variable and the three constant domains of the heavy chain, respectively.
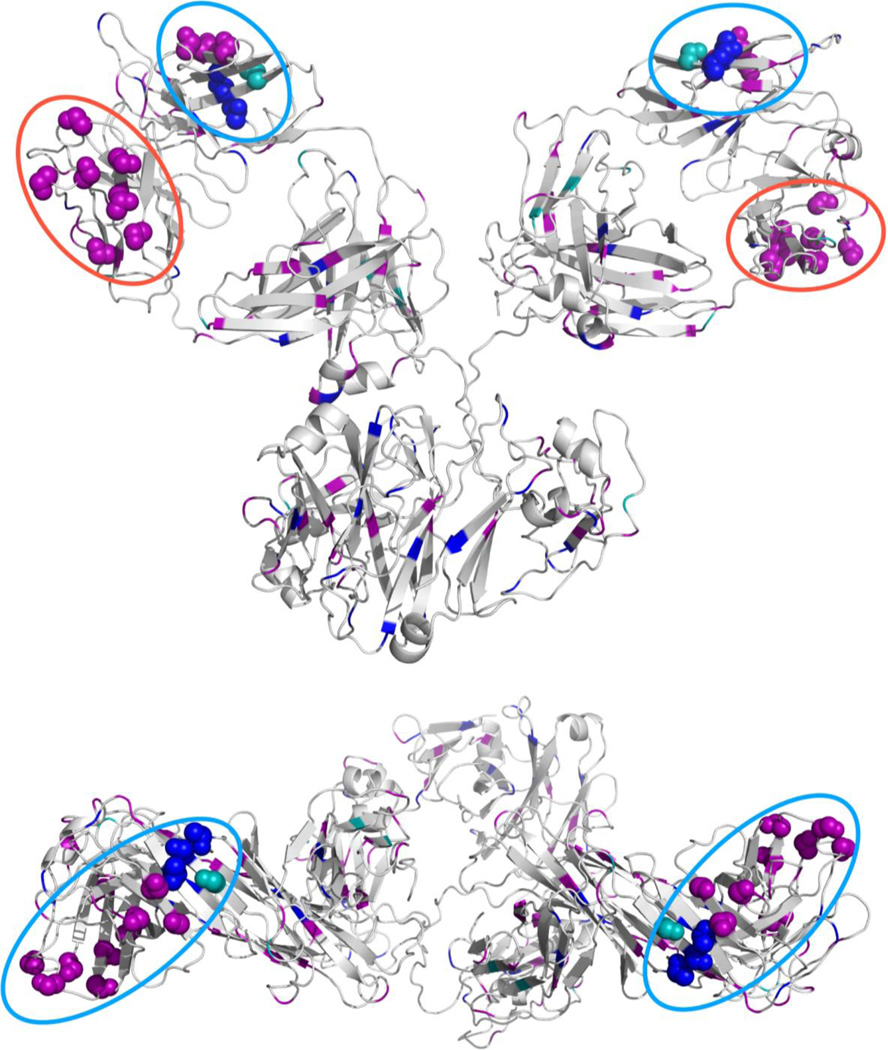
Greater structural insight is obtained by mapping the data in Table 2 onto an IgG homology model. A homology model was generated by the Swiss-Model workspace47,48 using an IgG crystal structure (PDB: 1IGY) as a template. As expected, the VL domain presents a large cluster of residues that undergo a decrease in labeling (Figure 3). Somewhat surprisingly, mapping the labeling data also reveals another potential interface on the VH domain, which also shows a clustering of residues undergoing a decrease in labeling. A color coded depiction of the different IgG domains in a space-filling model can be found in Figure S6 of the SI.
Figure 3.
Cartoon representations of IgG1 homology model. Side view (top) and top view (bottom). Spheres represent residues that are likely at the aggregate interface. Colors represent the magnitude of the reduction (Purple: >80%, Blue: 40–80%, and Teal: 10–40% reduction in labeling). The likely interfaces on the VL and VH domains are circled (VL: blue and VH: red).
The possible role that the VL and VH domains play in mediating IgG1 aggregates is further supported by the aggregation predictor tool Zyggregator (Tables S3 and S4 in the SI).49–52 When Zyggregator prediction data are overlaid onto IgG1’s structure (Figure S7 in the SI), there are four surface accessible regions in the protein that have a cluster of residues with a strong propensity to aggregate. Two of the four predicted sites are the VL and VH domains, which are implicated to be involved in aggregation by covalent labeling (Figure S7 in the SI). The other two predicted regions clearly have smaller surface areas, decreasing their likelihood of being true interfaces in the aggregates. It is interesting to note that previous studies are divided on which domains mediate aggregation processes in IgG’s. Some work suggests that typically the CH2 domain is the primary site of aggregation,2,53–55 while others suggest the variable regions in the Fab domain as the sites of aggregation.26,56–58 It is quite possible that the aggregation site varies from antibody to antibody.58 We feel our labeling results provide strong evidence for the role of the variable regions in the light and heavy chains in mediating aggregation in IgG1.
HGH
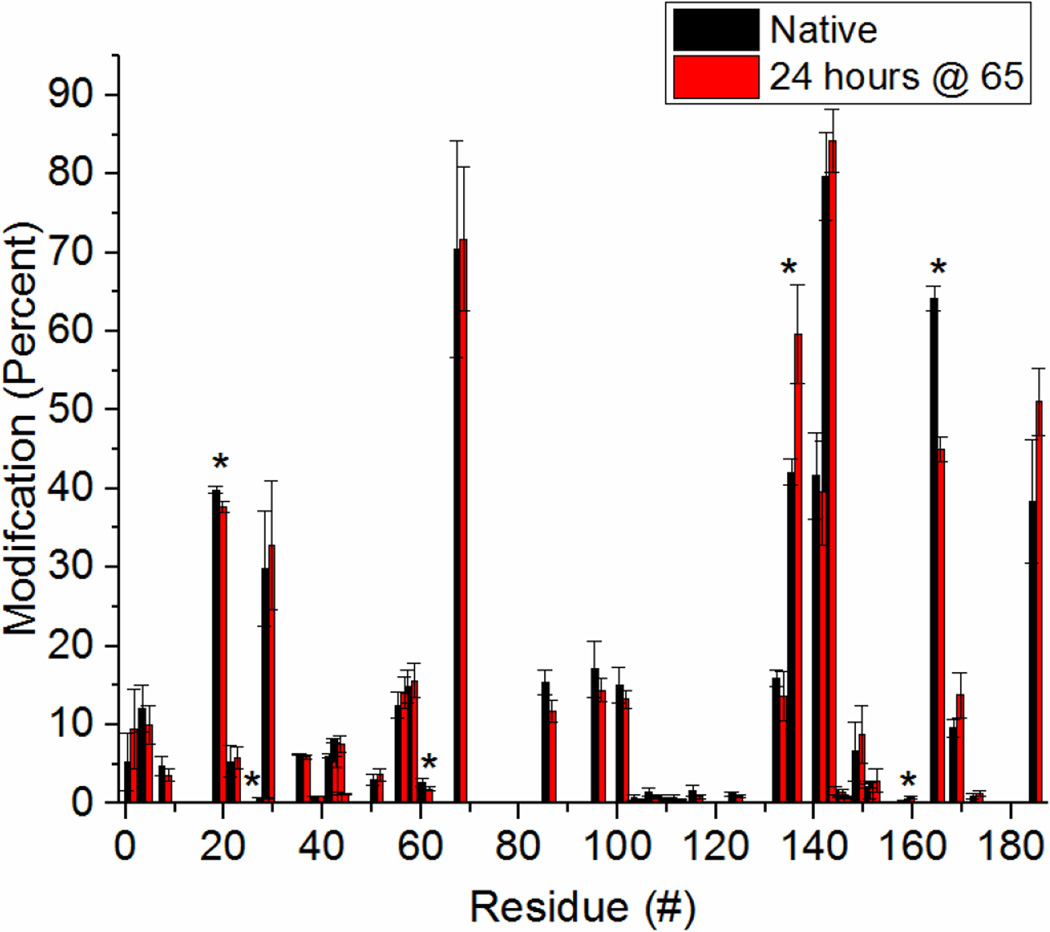
The results from IgG and β2m demonstrate that covalent labeling can identify proteins which have undergone severe structural perturbations. In order to test the technique’s ability to identify minor structural changes, we studied HGH. Labeling of this protein was performed before and after heating at 65 °C, a temperature that is 12 °C below its melting temperature.59 CD and intrinsic fluorescence spectra (Figures S8 and S9 in the SI) both show only minor structural perturbations after heating to this temperature. Upon DEPC labeling, 41 modification sites are identified (Table S5 in the SI) corresponding to over 20% of the protein. This amount of labeling ensures sufficient coverage of the protein’s structure. The labeling percentages of all the labeled sites for both native and heat-denatured HGH are summarized in Figure 4. When comparing the heat-denatured protein to the natively-structured protein, only six residues are found to undergo a significant change (p-value < 0.05) in labeling extent. They are His19, Thr28, Thr61, Thr136, Lys159, and Tyr165. Of these six, two residues undergo increased labeling upon heating (Thr136 and Lys159).
Figure 4.
Covlent labeling results for HGH before and after heating at 65°C for 24 hours. Asterisks (*) indicate residues that have undergone a statistically significant change. A difference was considered significant if the p-value, calculated by performing an unpaired T-test, was less than 0.05 (corresponding to a 95% confidence level at n=3).
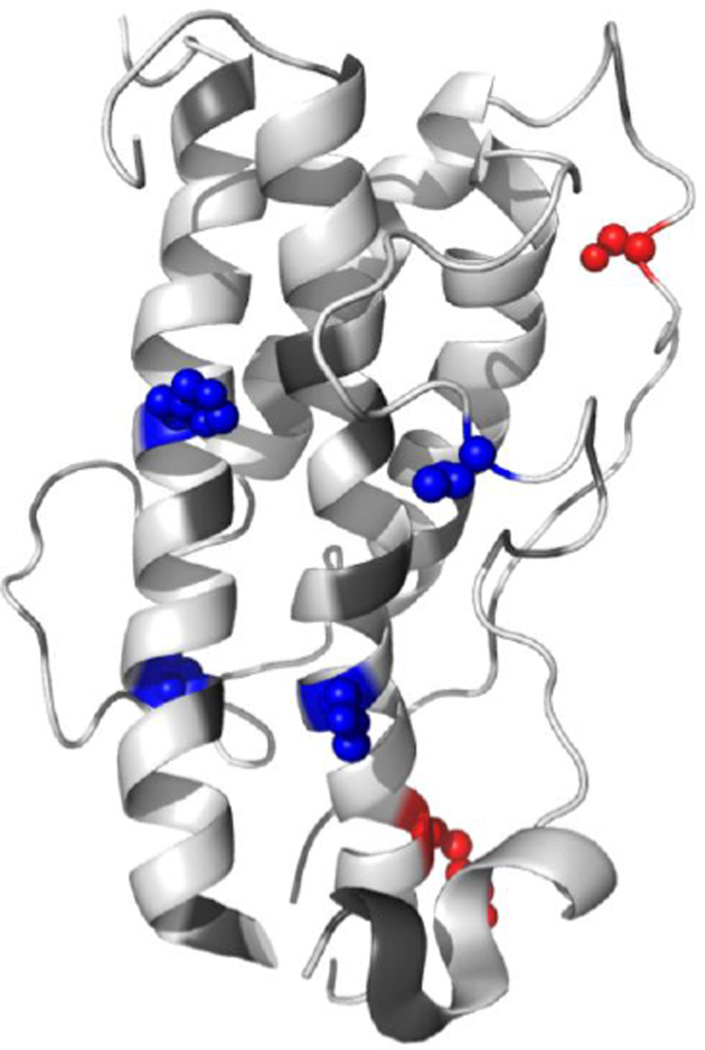
To understand the structural implications of these changes, we mapped the six residues on to a crystal structure of HGH (Figure 5, PDB: 1HGU). The two residues (Thr136 and Lys159) that undergo increased labeling after heating are on opposite ends of a long disordered region, signifying further melting of this region. The four residues (His19, Thr28, Thr61, and Tyr165) that undergo decreased labeling are clustered on the opposite face of the protein. While such clustering might suggest aggregation, DLS measurements (Figure S10 in the SI) indicate that the protein does not aggregate under these conditions. Instead, the DLS measurements reveal that the protein undergoes a slight compaction upon heating. Therefore, it is possible that the four residues undergo a decrease in labeling extent because they become less solvent exposed during this compaction process. Under denaturing conditions it has been demonstrated that HGH maintains a majority of its helical structure. Its loops, however, are known to become significantly more dynamic than the rest of the protein’s structure.60 Repositioning of these loops might cause Thr136 and Lys159 to become more solvent exposed, while at the same time causing His19, Thr28, Thr61, and Tyr165 to become less solvent exposed. Overall, these data for HGH suggest that DEPC labeling with MS detection is sensitive enough to detect minor structural perturbations.
Figure 5.
Summary of the covalent labeling results for HGH. Spheres represent residues that underwent significant changes in DEPC labeling after heating at 65 °C for 24 h (blue: decreased, red: increased). Changes in covalent labeling are mapped onto a crystal structure of HGH (PDB accession code: 1HGU).
CONCLUSIONS
We have used DEPC-based covalent labeling as a means to monitor the structure of therapeutic proteins. Through the study of three proteins, β2m, IgG, and HGH, we have shown that DEPC labeling is capable of identifying specific structural perturbations that occur upon exposing these proteins to common forced-degradation conditions. Because the label can probe up to 30% of the residues in a protein, this method provides a high degree of structural resolution relative to other covalent labeling reagents. DEPC labeling is particularly valuable for identifying interfacial residues in protein aggregates. For example, this technique was able to identify the variable domains of the light and heavy chain as the regions that mediate aggregation of IgG1 upon heating. DEPC labeling is also able to distinguish relatively minor perturbations in protein structure as illustrated by the experiments with HGH. Given the high effective resolution provided by DEPC labeling and the ease with which it can be performed relative to other MS-based techniques, we predict that this approach will be a powerful tool for studying therapeutic proteins.
Supplementary Material
Acknowledgments
The authors would like to thank Dr. Stephen Eyles for his help with the Thermo Scientific Orbitrap Fusion mass spectrometer, Dr. Jia Dong for her help with some of the SEC measurements, Bradley Duncan and Mark Hazelbaker for their assistance with the DLS measurements, and Joseph Tilitsky and Karan Hingorani for their help with the CD measurements. The authors would also like to thank Michael Knierman for his technical advice and helpful discussions, and Brandon Murphy for preliminary involvement in the work involving HGH. Acquisition of the Thermo Scientific Orbitrap Fusion was made possible by a grant from the National Institutes of Health S10OD010645. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
ABBREVIATIONS
- HRF
hydroxyl radical footprinting
- DEPC
diethylpyrocarbonate
- EDC
1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride
- GEE
glycine ethyl ester
- TCEP
tris(2-carboxyethyl)phosphine
- DTT
DL-dithiothreitol
Footnotes
ASSOCIATED CONTENT
Additional methods, biophysical techniques, example MS/MS spectra, TICs, and raw labeling data can be found in the supplemental information. These items can be found online free of charge at http://pubs.acs.org/.
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Aitken M. The Global Use of Medicines : Outlook Through 2015. Parsippany: 2013. [Google Scholar]
- 2.Fincke A, Winter J, Bunte T, Olbrich C. Eur. J. Pharm. Sci. 2014;62:148–160. doi: 10.1016/j.ejps.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 3.Schneider CK, Kalinke U. Nat. Biotechnol. 2008;26:985–990. doi: 10.1038/nbt0908-985. [DOI] [PubMed] [Google Scholar]
- 4.Ayoub D, Jabs W, Resemann A, Evers W, Evans C, Main L, Baessmann C, Wagner-Rousset E, Suckau D, Beck A. MAbs. 2013;5:699–710. doi: 10.4161/mabs.25423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Beck A, Diemer H, Ayoub D, Debaene F, Wagner-Rousset E, Carapito C, Van Dorsselaer A, Sanglier-Cianférani S. TrAC Trends Anal. Chem. 2013;48:81–95. [Google Scholar]
- 6.Katta V, Chait BT. Rapid Commun Mass Spectrom. 1991;5:214–217. doi: 10.1002/rcm.1290050415. [DOI] [PubMed] [Google Scholar]
- 7.Chowdhury SK, Katta V, Chait BT. J. Am. Chem. Soc. 1990;112:9012–9013. [Google Scholar]
- 8.Kaltashov IA, Eyles SJ. Mass Spectrom. Rev. 2002;21:37–71. doi: 10.1002/mas.10017. [DOI] [PubMed] [Google Scholar]
- 9.Engen JR, Smith DL. Anal. Chem. 2001;73:256 A–265 A. doi: 10.1021/ac012452f. [DOI] [PubMed] [Google Scholar]
- 10.Pirrone GF, Iacob RE, Engen JR. Anal. Chem. 2015;87:99–118. doi: 10.1021/ac5040242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck A, Debaene F, Diemer H, Wagner-Rousset E, Colas O, Van Dorsselaer A, Cianférani S. J. Mass Spectrom. 2015;50:285–297. doi: 10.1002/jms.3554. [DOI] [PubMed] [Google Scholar]
- 12.Pan LY, Salas-Solano O, Valliere-Douglass JF. Anal. Chem. 2015;87:5669–5676. doi: 10.1021/acs.analchem.5b00764. [DOI] [PubMed] [Google Scholar]
- 13.Wei H, Mo J, Tao L, Russell RJ, Tymiak AA, Chen G, Iacob RE, Engen JR. Drug Discov. Today. 2014;19:95–102. doi: 10.1016/j.drudis.2013.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Houde D, Arndt J, Domeier W, Berkowitz S, Engen JR. Anal. Chem. 2009;81:2644–2651. doi: 10.1021/ac802575y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leitner A, Walzthoeni T, Kahraman A, Herzog F, Rinner O, Beck M, Aebersold R. Mol. Cell. Proteomics. 2010;9:1634–1649. doi: 10.1074/mcp.R000001-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinz A. Mass Spectrom. Rev. 2006;25:663–682. doi: 10.1002/mas.20082. [DOI] [PubMed] [Google Scholar]
- 17.Back JW, de Jong L, Muijsers AO, de Koster CG. J. Mol. Biol. 2003;331:303–313. doi: 10.1016/s0022-2836(03)00721-6. [DOI] [PubMed] [Google Scholar]
- 18.Fancy D. Curr. Opin. Chem. Biol. 2000;4:28–33. doi: 10.1016/s1367-5931(99)00047-2. [DOI] [PubMed] [Google Scholar]
- 19.Kluger R, Alagic A. Bioorg. Chem. 2004;32:451–472. doi: 10.1016/j.bioorg.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Peter JF, Tomer KB. Anal. Chem. 2001;73:4012–4019. doi: 10.1021/ac010258n. [DOI] [PubMed] [Google Scholar]
- 21.Hambly DM, Gross ML. J. Am. Soc. Mass Spectrom. 2005;16:2057–2063. doi: 10.1016/j.jasms.2005.09.008. [DOI] [PubMed] [Google Scholar]
- 22.Hambly D, Gross M. Int. J. Mass Spectrom. 2007;259:124–129. [Google Scholar]
- 23.Xu G, Chance MR. Chem. Rev. 2007;107:3514–3543. doi: 10.1021/cr0682047. [DOI] [PubMed] [Google Scholar]
- 24.Konermann L, Stocks BB, Pan Y, Tong X. Mass Spectrom. Rev. 2010;29:651–667. doi: 10.1002/mas.20256. [DOI] [PubMed] [Google Scholar]
- 25.Watson C, Sharp JS. AAPS J. 2012;14:206–217. doi: 10.1208/s12248-012-9336-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Deperalta G, Alvarez M, Bechtel C, Dong K, McDonald R, Ling V. MAbs. 2012;5:86–101. doi: 10.4161/mabs.22964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mendoza VL, Vachet RW. Mass Spectrom. Rev. 2009;28:785–815. doi: 10.1002/mas.20203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kaur P, Tomechko SE, Kiselar J, Shi W, Deperalta G, Wecksler AT, Gokulrangan G, Ling V, Chance MR. MAbs. 2015;7:540–552. doi: 10.1080/19420862.2015.1023683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zhou Y, Vachet RW. J. Am. Soc. Mass Spectrom. 2012;23:708–717. doi: 10.1007/s13361-011-0332-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mendoza VL, Vachet RW. Anal. Chem. 2008;80:2895–2904. doi: 10.1021/ac701999b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Srikanth R, Mendoza VL, Bridgewater JD, Zhang G, Vachet RW. Biochemistry. 2009;48:9871–9881. doi: 10.1021/bi901172y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mendoza VL, Antwi K, Barón-Rodríguez Ma, Blanco C, Vachet RW. Biochemistry. 2010;49:1522–1532. doi: 10.1021/bi901748h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mendoza VL, Barón-Rodríguez MA, Blanco C, Vachet RW. Biochemistry. 2011;50:6711–6722. doi: 10.1021/bi2004894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhou Y, Vachet RW. J. Am. Soc. Mass Spectrom. 2012;23:899–907. doi: 10.1007/s13361-012-0349-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chambers MC, Maclean B, Burke R, Amodei D, Ruderman DL, Neumann S, Gatto L, Fischer B, Pratt B, Egertson J, Hoff K, Kessner D, Tasman N, Shulman N, Frewen B, Baker TA, Brusniak M-Y, Paulse C, Creasy D, Flashner L, Kani K, Moulding C, Seymour SL, Nuwaysir LM, Lefebvre B, Kuhlmann F, Roark J, Rainer P, Detlev S, Hemenway T, Huhmer A, Langridge J, Connolly B, Chadick T, Holly K, Eckels J, Deutsch EW, Moritz RL, Katz JE, Agus DB, MacCoss M, Tabb DL, Mallick P. Nat. Biotechnol. 2012;30:918–920. doi: 10.1038/nbt.2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaudel M, Barsnes H, Berven FS, Sickmann A, Martens L. Proteomics. 2011;11:996–999. doi: 10.1002/pmic.201000595. [DOI] [PubMed] [Google Scholar]
- 37.Craig R, Beavis RC. Rapid Commun. Mass Spectrom. 2003;17:2310–2316. doi: 10.1002/rcm.1198. [DOI] [PubMed] [Google Scholar]
- 38.Craig R, Beavis RC. Bioinformatics. 2004;20:1466–1467. doi: 10.1093/bioinformatics/bth092. [DOI] [PubMed] [Google Scholar]
- 39.Dorfer V, Pichler P, Stranzl T, Stadlmann J, Taus T, Winkler S, Mechtler K. J. Proteome Res. 2014;13:3679–3684. doi: 10.1021/pr500202e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim S, Pevzner PA. Nat. Commun. 2014;5:5277–5286. doi: 10.1038/ncomms6277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. J. Proteome Res. 2004;3:958–964. doi: 10.1021/pr0499491. [DOI] [PubMed] [Google Scholar]
- 42.Eng JK, Jahan TA, Hoopmann MR. Proteomics. 2013;13:22–24. doi: 10.1002/pmic.201200439. [DOI] [PubMed] [Google Scholar]
- 43.Vaudel M, Burkhart JM, Zahedi RP, Oveland E, Berven FS, Sickmann A, Martens L, Barsnes H. Nat. Biotechnol. 2015;33:22–24. doi: 10.1038/nbt.3109. [DOI] [PubMed] [Google Scholar]
- 44.Pluskal T, Castillo S, Villar-Briones A, Orešič M. BMC Bioinformatics. 2010;11:395–406. doi: 10.1186/1471-2105-11-395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wiedemann C, Bellstedt P, Görlach M. Bioinformatics. 2013;29:1750–1757. doi: 10.1093/bioinformatics/btt278. [DOI] [PubMed] [Google Scholar]
- 46.Madden DR, Gorga JC, Strominger JL, Wiley DC. Cell. 1992;70:1035–1048. doi: 10.1016/0092-8674(92)90252-8. [DOI] [PubMed] [Google Scholar]
- 47.Bordoli L, Kiefer F, Arnold K, Benkert P, Battey J, Schwede T. Nat. Protoc. 2009;4:1–13. doi: 10.1038/nprot.2008.197. [DOI] [PubMed] [Google Scholar]
- 48.Arnold K, Bordoli L, Kopp J, Schwede T. Bioinformatics. 2006;22:195–201. doi: 10.1093/bioinformatics/bti770. [DOI] [PubMed] [Google Scholar]
- 49.Pawar AP, DuBay KF, Zurdo J, Chiti F, Vendruscolo M, Dobson CM. J. Mol. Biol. 2005;350:379–392. doi: 10.1016/j.jmb.2005.04.016. [DOI] [PubMed] [Google Scholar]
- 50.DuBay KF, Pawar AP, Chiti F, Zurdo J, Dobson CM, Vendruscolo M. J. Mol. Biol. 2004;341:1317–1326. doi: 10.1016/j.jmb.2004.06.043. [DOI] [PubMed] [Google Scholar]
- 51.Tartaglia GG, Pawar AP, Campioni S, Dobson CM, Chiti F, Vendruscolo M. J. Mol. Biol. 2008;380:425–436. doi: 10.1016/j.jmb.2008.05.013. [DOI] [PubMed] [Google Scholar]
- 52.Tartaglia GG, Vendruscolo M. Chem. Soc. Rev. 2008;37:1395–1401. doi: 10.1039/b706784b. [DOI] [PubMed] [Google Scholar]
- 53.Andersen CB, Manno M, Rischel C, Thórólfsson M, Martorana V. Protein Sci. 2010;19:279–290. doi: 10.1002/pro.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Van Buren N, Rehder D, Gadgil H, Matsumura M, Jacob J. J. Pharm. Sci. 2009;98:3013–3030. doi: 10.1002/jps.21514. [DOI] [PubMed] [Google Scholar]
- 55.Ito T, Tsumoto K. Protein Sci. 2013;22:1542–1551. doi: 10.1002/pro.2340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Sahin E, Grillo AO, Perkins MD, Roberts CJ. J. Pharm. Sci. 2010;99:4830–4848. doi: 10.1002/jps.22198. [DOI] [PubMed] [Google Scholar]
- 57.Brummitt RK, Nesta DP, Chang L, Chase SF, Laue TM, Roberts CJ. J. Pharm. Sci. 2011;100:2087–2103. doi: 10.1002/jps.22448. [DOI] [PubMed] [Google Scholar]
- 58.Wu H, Kroe-Barrett R, Singh S, Robinson AS, Roberts CJ. FEBS Lett. 2014;588:936–941. doi: 10.1016/j.febslet.2014.01.051. [DOI] [PubMed] [Google Scholar]
- 59.Mulinacci F, A.H. Capelle M, Gurny R, F. Drake A, Arvinte T. J. Pharm. Sci. 2011;100:451–463. doi: 10.1002/jps.22293. [DOI] [PubMed] [Google Scholar]
- 60.Kasimova MR, Kristensen SM, Howe PWA, Christensen T, Matthiesen F, Petersen J, Sørensen HH, Led JJ. J. Mol. Biol. 2002;318:679–695. doi: 10.1016/S0022-2836(02)00137-7. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.