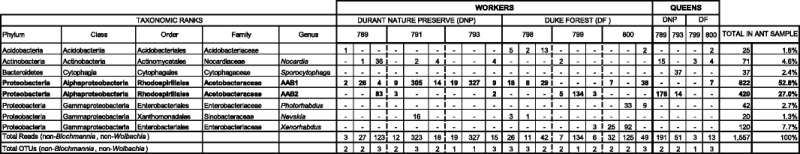
Table 1.
Abundance (read count) of Bacterial OTUs detected in Camponotus chromaiodes. Read counts are based on 16S amplicon sequencing. “-” indicates zero reads detected. Endosymbiont reads (Blochmannia and Wolbachia) have been removed from the read counts shown. Only OTUs comprising >1 % of total reads are shown. Each worker sample represents a pool of 3–5 surface-sterilized whole gasters. Colonies are demarcated by a dashed line line and listed with a three digit identifier. The total % contribution of each OTU (right-most column) reflects the total read count of that OTU, among the 1,557 non-Blochmannia, non-Wolbachia reads for all 22 ant samples, after excluding OTUs <1 %. AAB1 & 2 are displayed in boldface