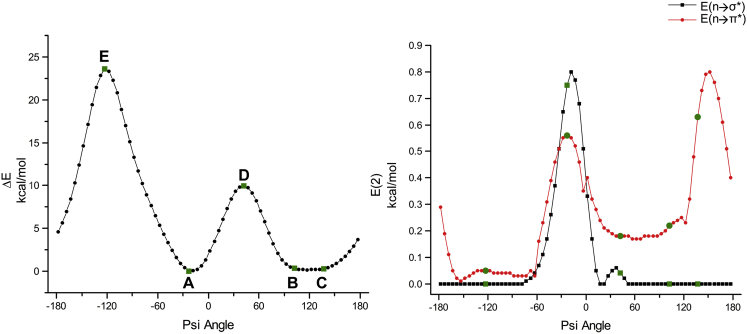
Figure 3.
Left: potential energy profile generated from the single-point energy calculation of the model compound Ace-Pro-NMe as a function of ψ. The initial conformation corresponds to the Pro-94 residue of the experimentally determined protein structure cytochrome peroxidase (PDB: 4CVI) and the ψ angle of Pro-94 in this structure is −22.9°. Starting from this conformation, the ψ angle was varied in steps of 5° and single-point energy calculations were carried out as described in Materials and Methods on 72 conformations. Each point in the plot represents the relative energy with respect to the initial conformation. (A) This conformation corresponds to the experimentally determined initial conformation and is the energy minimum. (B and C) Conformations that are energetically closer to (A). (D and E) Two peaks in the profile. Right: the second-order perturbation energy profile was plotted for all conformations of Ace-Pro-NMe for different ψ values, and NBO calculations for each conformation were carried out as described in Materials and Methods. The energy profiles shown in filled squares and filled circles represent N-H…N H-bond and n → π∗ interactions, respectively. To see this figure in color, go online.