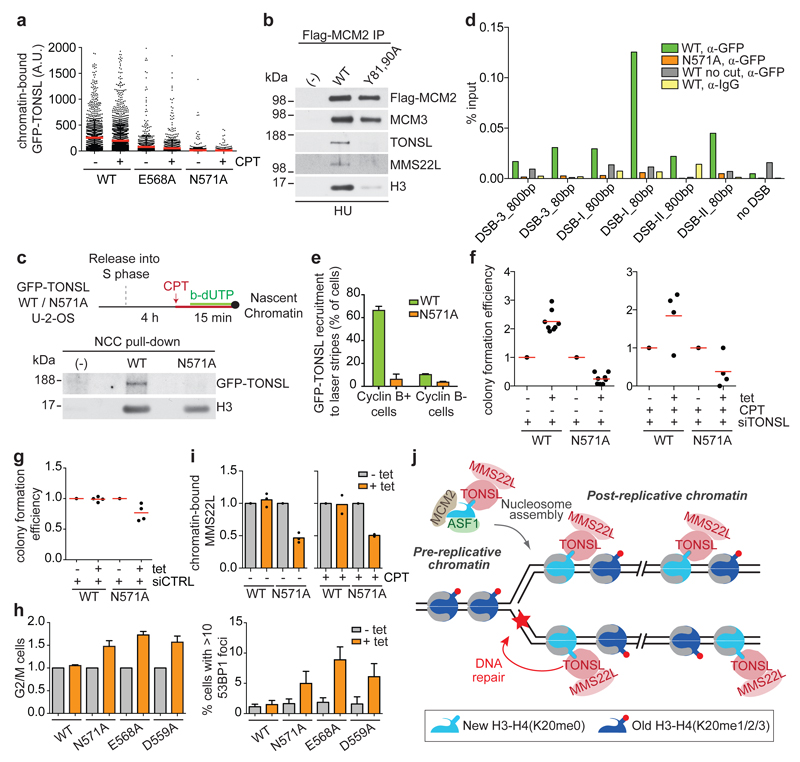
Figure 4. H4K20me0 recognition is required for TONSL accumulation at DNA repair sites and genome stability.
a, Chromatin-binding of GFP-TONSL in CPT treated S-phase cells. Error bar, SD; from left, n=1461, 2631, 1245, 1764, 2116, 3178. b, Co-immunoprecipitation of TONSL-MMS22L with Flag-HA-MCM2 WT or histone binding mutant (Y81A, Y90A)7 from chromatin after HU treatment. c, NCC analysis of GFP-TONSL recruitment to replication forks in CPT treated cells. (-), no biotin-dUTP. d, ChIP-qPCR analysis of GFP-TONSL recruitment to site-specific DSBs induced by AsiSI29. See Extended data Fig. 8a for additional controls. e, GFP-TONSL recruitment to laser-induced DNA lesions (error bars, SD; n=3; total cells counted, 210 (WT) and 252 (N571)). f, g, Colony formation upon GFP-TONSL induction (+tet) in siRNA and CPT treated cells. h, Cell cycle and 53BP1 foci analysed by microscopy. (left) % G2/M cells shown relative to non-induced. Error bars, SD, n= 4 (left), 5 (right). i, Chromatin-bound MMS22L analysed as in Fig. 3e. Mean with individual data points are shown (n=3 (untreated), 2 (CPT)), see Extended Data Fig. 8i for western blots. j, TONSL-MMS22L identifies post-replicative chromatin by binding H4K20me0 on new histones, directing TONSL-MMS22L genome surveillance function to DNA having a sister chromatid. Data are representative of 3 (a), 2 (b-d, f-right, g), and 4 (f-left) independent experiments. Protein inputs, Extended Data Fig. 9e, f.