Abstract
EGFR mutation testing is now well established as a means of selecting the optimal first‐line therapy for patients with advanced non‐small‐cell lung cancer (NSCLC). However, deciding on the correct treatment for EGFR wild‐type NSCLC remains a challenge. EGFR fluorescence in‐situ hybridization (FISH) testing of gene copy number has been a promising marker, but has provided mixed results despite attempts to standardize the reading and scoring process. The novel ReadMax reading and scoring system focuses on the most aberrant cells, to identify oncogene addiction, rather than taking a representative reading as in the Colorado method. The methodology was developed using historical samples from the TRUST and MERIT studies, followed by re‐reading of the samples from the SATURN trial. Analysis of samples using the ReadMax methodology revealed that progression‐free survival (PFS) and overall survival (OS) were improved in patients with ReadMax FISH‐positive (RM FISH+) tumours, compared with those whose tumours were not RM FISH+: PFS hazard ratios (HRs) were 0.52 for RM FISH+ versus 0.93 for not RM FISH+; OS HRs were 0.69 and 0.92, respectively. For PFS, HR for RM FISH+ versus not RM FISH+ in the SATURN erlotinib group was 0.53 (p = 0.003). The PFS and OS results were also similar in the EGFR wild‐type population (PFS HRs were 0.63 and 0.96; OS HRs were 0.61 and 0.84, respectively), although amplification of the EGFR gene in patients with EGFR wild‐type disease was not found to be predictive of treatment outcomes, which was unexpected but not unprecedented. KRAS status was not found to affect outcomes. Further experience is required to refine the ReadMax methodology and fully establish its validity and robustness. In conclusion, the ReadMax scoring system to identify patients with EGFR FISH‐positive NSCLC is a promising technique, which could improve treatment options and outcomes for patients with advanced NSCLC, in particular for EGFR wild‐type tumours.
Keywords: non‐small‐cell lung cancer, EGFR, biomarkers, gene expression, FISH, ReadMax
Introduction
EGFR mutation testing is now well established as a means of selecting the optimal first‐line therapy for patients with advanced non‐small‐cell lung cancer (NSCLC). However, deciding on the correct treatment choice for EGFR wild‐type NSCLC remains a challenge, as no adequate predictive marker has been found to date. Given the importance of EGFR as a target, especially for the tyrosine kinase inhibitors (TKIs), it remains a marker of interest in NSCLC. Fluorescence in‐situ hybridization (FISH) testing of EGFR gene copy number (GCN) was identified as a biomarker for targeted therapy in advanced NSCLC, and early studies in advanced NSCLC found evidence of a predictive relationship between EGFR FISH status and outcomes with EGFR TKIs 1, 2, 3. However, EGFR FISH testing according to the 2005 Colorado system 4 has provided mixed results across different clinical trials.
In order to improve standardization of reading and scoring across laboratories with this system, a revised version of the Colorado testing guidance was issued 4 and employed in the phase III SATURN study of erlotinib in the maintenance setting. However, despite differences in progression‐free survival (PFS) between patients whose tumours were Colorado FISH‐positive (hazard ratio [HR] = 0.68; 95% confidence interval [CI] 0.51–0.90; p = 0.0068) and those with FISH‐negative tumours (HR = 0.81; 95% CI 0.62–1.07; p = 0.13), the study found no significant association between Colorado FISH status and outcomes with erlotinib (interaction p = 0.35) 5. In the EGFR wild‐type population of SATURN, patients with Colorado FISH‐positive disease performed slightly worse than those with Colorado FISH‐negative disease (see supplementary material, Figure S1), which was contrary to expectations. Still, it appears that there is a link between EGFR gene amplification and the presence of EGFR mutations 6, which underlines the predictive potential of EGFR FISH.
In view of this spectrum of mixed results, the established FISH reading and scoring approach was put into question. The original reading strategy aims to capture a representative subset of cells and, when scoring the FISH status, the estimated prevalence of aberrant cells in the sample is used (Figure 1A). This approach faces difficulties when assessing heterogeneous lung tumour samples with focal spots of amplification and randomly distributed, isolated, highly aberrant cells; the representative reading approach would lead to ‘washing out’ of aberrance. The main idea of the revised approach was to determine the existence and pattern of aberrance in the sample, rather than to quantify the proportion of aberrance, ie changing from a quantitative to a qualitative assessment (Figure 1B). This could also help address the problem of high variability within the lung tumour samples and allows for the assessment of samples with small numbers of viable tumour cells. The existence of aberrant cells is likely to signify oncogene addiction of the tumour to the EGFR pathway. The most aberrant cells might drive the tumour biology, and therefore, may directly relate to the likelihood of response to EGFR TKIs. A similar approach is used currently in human epithelial growth factor receptor 2 (HER2) testing in breast cancer, where the most aberrant cells are read as a priority.
Figure 1.
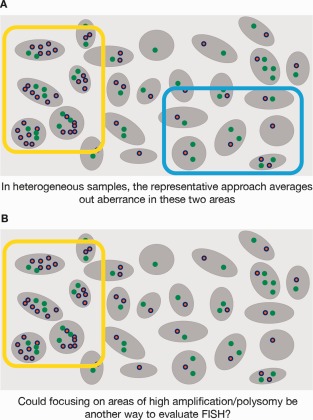
Representative versus maximization approach in assessing GCN.
First tests of this ‘maximization’ idea were retrospectively based on existing representative FISH readings from a phase II, single‐arm, gene expression profiling study of erlotinib in NSCLC (MERIT) 7 and a phase IV, open‐label study of erlotinib in the clinical practice setting (TRUST) 8 (40 or 100 cells, depending on laboratory; see supplementary material for more details). The ten cells with highest GCN were selected and samples were classified based on the mean GCN in these ten cells (top ten). Existing data from 83 patients from the MERIT study, 208 patients from TRUST‐1 and 56 patients from TRUST‐2 were re‐categorized. Categorization of the samples from MERIT and TRUST based on this surrogate measure demonstrated a clear difference in terms of treatment outcome for ‘top ten’ FISH‐positive versus ‘top ten’ FISH‐negative NSCLC: for MERIT, an HR of 0.60 was observed for PFS (95% CI 0.41–1.00; p = 0.05). These data emphasized the importance of adequate scoring approaches and warranted a deeper exploration of using ‘maximization of aberrance’ as the principal reading strategy, associated with a systematic exploration of elaborate scoring approaches to quantify different patterns of aberrance (see supplementary material for more details).
These preliminary results of the simple piggy‐back ‘top ten’ approach merited exploration of the potential of a maximization reading strategy. In two feasibility studies, the maximizing reading scheme of 20 cells was successfully tested by pathologists (see more details in supplementary material). Finally, an exploratory study was set up based on re‐examining existing SATURN and TITAN FISH slides to address the following goals: establish a new maximizing reading strategy (ReadMax) to capture the 20 most aberrant cells, with potential gains in reader‐to‐reader variability and in pathologist reading time; find and quantify different patterns of aberrance (eg amplification and variants of polysomy); systematically evaluate different advanced scoring approaches by diagnostic criteria; explore the predictive power of different FISH patterns regarding treatment with erlotinib; and specifically explore predictiveness for the EGFR wild‐type population.
Materials and methods
Clinical studies and FISH re‐reading
The SATURN study was a phase III, global, placebo‐controlled study initiated to evaluate erlotinib as maintenance therapy following standard first‐line chemotherapy regimens in patients with advanced NSCLC. A total of 889 patients were eligible for entry into the maintenance phase of the study and tumour sampling for biomarker analysis was mandatory. Full methodology has been published previously, along with both clinical and biomarker analyses 5, 9. The TITAN study was a phase III study that enrolled patients who were ineligible to enter the SATURN study after the initial chemotherapy run‐in phase, ie those who had progressed within the initial four cycles of chemotherapy. These 424 patients were randomized 1:1 to receive second‐line erlotinib or chemotherapy (either docetaxel or pemetrexed, at the investigators' discretion). As with SATURN, tumour sampling was mandatory in TITAN, and full methodology and results have been reported 10.
Out of the total of 485 and 242 stored FISH slides from SATURN and TITAN, respectively, 201 (41%) and 171 (70%) met the quality requirements for re‐reading (ie signals were not faded due to the time since original processing). The subset of samples eligible by quality was found to be representative of the overall study population in terms of demographics and primary morbidity parameters.
All studies included in this analysis were carried out in accordance with the Declaration of Helsinki and Good Clinical Practice guidelines. Protocols were approved by local ethics committees at each investigating centre, and all enrolled patients gave informed written consent to participate in the studies and provide tumour samples for biomarker analysis.
ReadMax instruction and raw reading result
Beginning with stained slides, the pathologist scanned the tumour tissue for areas of highest EGFR GCN. Selecting a total of 20 cells in four areas (5 cells/area) with the highest EGFR GCN, they then recorded the count of EGFR and chromosome 7 centromere (CEP7) for each of these cells. This produced a table of 40 numbers, with matched pairs of EGFR and CEP7 values for each of the 20 cells. To ensure reliability, it was essential that the pathologist thoroughly scanned all parts of the tumour sample for areas with the highest EGFR GCN.
Basic quantification of raw reading result
The raw readings were categorized into ten descriptive quantities: five features relate to the GCN of the target gene, counting the nuclei with a particular signal count (1–2, 3–4, 5–6, 7–12 and >12); and five features count nuclei according to the relation of target to centromere signals (counting nuclei where target minus centromere is ≤0, 1, 2–4 and >4; the fifth feature is the number of nuclei with ratio >2). Altogether, this defines ten numbers, which should characterize the FISH reading.
Systematic development of scoring features with multivariate analysis
The next step required extraction of features that could describe the different patterns of EGFR GCN aberration using a ‘bottom‐up’ approach, with the data determining which different FISH patterns were discernible in this disease population (based on the ten numbers characterizing the FISH reading). Several multivariate approaches were used to extract scoring candidates from the data. Some were discounted because of redundancy and poor diagnostic stability. Approaches that showed promising properties were: the relevant dimensions of the actual FISH patterns observed (by different approaches of factor analysis) based on FISH data only; scoring derived through pattern recognition methods by comparing the FISH patterns of mutated tumours versus wild‐type tumours; scoring derived by pattern recognition methods comparing the FISH pattern of long survivors with early progressors in a subset of the erlotinib arm; and scorings derived from regression approaches involving the PFS endpoint.
Selecting scorings with favourable diagnostic properties
Scoring approaches underwent diagnostic stability analysis and were assessed for their robustness regarding the heterogeneity of raw readings. Scorings selected by diagnostic quality were transformed from simple implicit cutoffs of 0–1 to so‐called ‘propensities’ (derived by statistical re‐sampling techniques; see supplementary material), which express, on a 0–100% scale, the propensity of a FISH reading to lie above the cutoff of the scoring. Heterogeneous tumours may lead to heterogeneous FISH readings, and this may result in intermediate propensities, ie far from the values 0 or 100.
The practicability and reproducibility of the ReadMax strategy was assessed in a ring study including two laboratories with two readers each. See supplementary material for more details.
Endpoints for assessing the predictive value
Endpoints assessed were PFS, overall survival (OS) and disease control rate (DCR) in all comers and in patients with EGFR wild‐type NSCLC.
Statistical analyses
PFS and OS were analysed using Kaplan–Meier methodology, with HRs and 95% CIs calculated using Cox regression analysis. Time‐to‐event endpoints were measured from randomization. Overall response (complete response + partial response) and DCR (complete response + partial response + stable disease) were calculated using Response Evaluation Criteria in Solid Tumors (RECIST) version 1.0 and were expressed as odds ratios with 95% CIs, using logistic regression.
Results
Diagnostic evaluation
Defined patterns of aberration
Discernible patterns of EGFR GCN aberration were found, which correlated with previously described patterns such as amplification and polysomy; however, they also showed other features that elude description by standard terms. Four patterns of aberration were defined. EGFR_MULT was derived from multivariate factor analysis based on FISH data only; this was characterized by high EGFR GCN and low CEP7 counts (ie it resembles classical ‘amplification'). HI_POLY was derived by comparing the FISH pattern from early‐progressing patients or those with long PFS using pattern recognition methods; this category bears a resemblance to high polysomy (ie high EGFR GCN accompanied by high CEP7 counts). FACS_MUT was derived by comparing the FISH pattern of tumours harbouring activating EGFR mutations with FISH patterns found in wild‐type tumours, using pattern recognition methods. In this way, wild‐type samples with FISH patterns resembling mutation‐positive FISH patterns, ie facsimile mutations, can be identified. POLY_F2 was derived from multivariate factor analysis; it scores high for a specific polysomial pattern with medium high counts of EGFR and CEP7.
The possibility for an optimistic bias when extracting these aberration patterns must be considered. For the patterns EGFR_MULT, FACS_MUT and POLY_F2 this can be excluded, since the extraction was based on the FISH readings and no outcome data were used. These scorings can, therefore, be considered to describe the dimensions of FISH aberrance patterns present in this NSCLC population and there is no a priori interaction with treatment outcome.
Since the pattern HI_POLY was derived using the PFS outcome in the erlotinib arm, an overoptimistic bias could be possible in this group. In order to minimize this potential bias, the pattern extraction was based on a randomly selected subgroup. However, since the endpoint results for HI_POLY were indeed favourable, an additional adjustment run was done in order to correct for this potential bias. Based on 1000 random permutations, the raw results of HI_POLY were corrected with a HR for erlotinib versus placebo for the HI_POLY‐positive group of 0.44 (p = 0.001; adjusted p = 0.016) and a HR for HI_POLY positive versus HI_POLY negative for the erlotinib group of 0.46 (p = 0.0; adjusted p = 0.006). The interaction HR was 0.43 (p = 0.007; adjusted p = 0.017). These results provide reassurance that the potential bias of HI_POLY is negligible.
Regarding cutoffs, the four scorings derived from multivariate procedures possess an implicit cutoff, hence no additional cutoff search was needed for these. These implicit cutoffs are based on the FISH pattern only, requiring no reference to the endpoints. The implicit cutoff is zero for scorings evolving from factor analysis (ie EGFR_MULT, POLY_F2), as the scores are construed to give a mean of zero in the given population. Similarly, scorings derived from pattern recognition methods (ie FACS_MUT, HI_POLY) are construed to provide a classifier to separate two groups based on the given FISH pattern; this represents the implicit cutoff.
The four categories were each based on a set of coefficients, so the spread of these coefficients was assessed based on statistical re‐sampling techniques and testing through propensities, in order to establish the diagnostic stability of these scoring approaches (see supplementary material for more detail).
For the predictive analysis reported here, the following cutoffs for propensities of each of the four categories were used: ≥65% was positive, ≤35% was negative and >35% to <65% was equivocal. For each of the four categories, the predictive value regarding the endpoints was individually investigated by comparing the performance of positives versus negatives, ie leaving out the equivocal cases. It is obvious that each of the aberrance features possess some predictive value regarding the endpoints PFS, OS and DCR, partially achieving statistical significance and demonstrating consistent effects even when significance was not reached due to low sample size and ensuing wide confidence intervals.
In order to provide a provisional rule on how these four features could be combined to achieve an overall assessment, it was observed that EGFR_MULT was not predictive in EGFR wild‐type patients. Therefore, the following decision rule for defining the ReadMax status as positive (RM FISH‐positive) was established: samples where EGFR mutation status was unknown were classed as RM FISH‐positive if the sample was either EGFR_MULT‐positive or two out of three of HI_POLY, FACS_MUT and POLY_F2 were positive. Known EGFR wild‐type samples were classed as RM FISH‐positive if two out of three of HI_POLY, FACS_MUT and POLY_F2 were positive, without regard to EGFR_MULT (Table 1).
Table 1.
Interpretation of ReadMax based on aberration patterns and EGFR mutation status
| EGFR mutation status | HI_POLY/FACS_MUT/POLY_F2 | EGFR_MULT | ReadMax outcome |
|---|---|---|---|
| Unknown status | 2/3 or 3/3 positive | Any | Positive |
| All not positive or 1/3 positive | Positive | Positive | |
| All not positive or 1/3 positive | Not positive | Not positive | |
| Known wild type | 2/3 or 3/3 positive | N/A | Positive |
| All not positive or 1/3 positive | N/A | Not positive |
As the scoring approach for the four aberrance features allows for equivocal results, those not positive according to the combined RM definition could involve equivocal cases from the four patterns. Therefore, a clear definition of what is RM FISH‐negative is difficult to establish. In order not to preclude a definition of clear negativity in the future, this report refers to the category of ‘not RM FISH‐positive’ instead, which includes both cases that are clearly negative and those that are equivocal.
Predictive endpoint analysis
From the total of 201 re‐read samples from SATURN, there were 24 EGFR mutation positive, 131 EGFR wild type and 46 EGFR unknown. Around one‐third of patients with EGFR wild‐type disease were classed as RM FISH‐positive, while the percentage of patients with RM FISH‐positive status in the EGFR mutation‐positive group was much higher, in accordance with previous observations (Figure 2).
Figure 2.
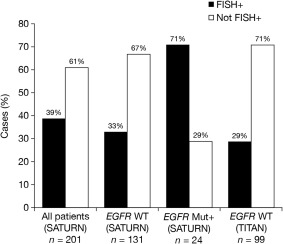
ReadMax results in SATURN and TITAN samples.
PFS, OS and DCR were all improved in patients with RM FISH‐positive tumours, compared with those whose tumours were not RM FISH‐positive (Figure 3, Table 2; see supplementary material, Figure S2 for outcomes by specific score). The PFS and OS results were also similar in the EGFR wild‐type population (Figure 4, Table 2; PFS: RM+ HR = 0.63, not RM+ HR = 0.96; OS: RM+ HR = 0.61, not RM+ HR = 0.84). Amplification of the EGFR gene (EGFR_MULT) in patients with EGFR wild‐type disease was not found to be predictive of treatment outcomes (supplementary material, Figure S3; PFS: EGFR_MULT+ HR = 0.86, EGFR_MULT− HR=0.85, interaction = 1.02, p > 0.9), which was unexpected.
Figure 3.
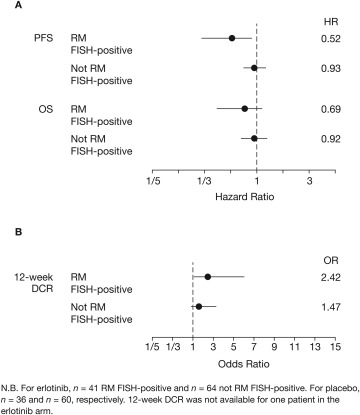
Forest plots of (A) PFS and OS; and (B) 12‐week DCR according to ReadMax category for all‐comers in SATURN (RM set).
Table 2.
Comparison of PFS by original Colorado scoring versus ReadMax scoring in the SATURN study (ReadMax population)
| HR (p value) | |||
|---|---|---|---|
| Comparison | Colorado scoring | ReadMax scoring | |
| All patients with data (n = 201) | Erlotinib FISH+ vs erlotinib not FISH+ | 0.73 (p = 0.136) | 0.53 (p = 0.003) |
| Erlotinib FISH+ vs placebo FISH+ | 0.67 (p = 0.04) | 0.52 (p = 0.004) | |
| FISH‐positive (%) | 55 | 38 | |
| EGFR WT subgroup (n = 131) | Erlotinib FISH+ vs erlotinib not FISH+ | 1.20 (p = 0.48) | 0.65 (p = 0.143) |
| Erlotinib FISH+ vs placebo FISH+ | 1.02 (p = 0.94) | 0.63 (p = 0.185) | |
| FISH‐positive (%) | 51 | 27 | |
WT, wild‐type
Figure 4.
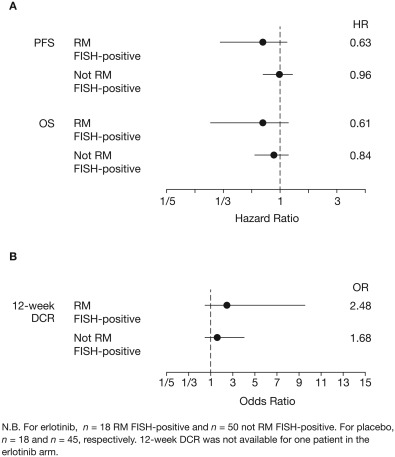
Forest plots of (A) PFS and OS; and (B) 12‐week DCR according to ReadMax category for the EGFR wild‐type population in SATURN (RM set).
Of the 131 EGFR wild types in this study, 102 were confirmed also to be KRAS wild type. Analysis of this group of 102 ‘double wild type’ patients found similar results to the overall EGFR wild‐type group of 131: RM+ HR = 0.62 for erlotinib versus placebo, p = 0.294; HR = 0.55 for RM+ versus not RM+ in the erlotinib group only, p = 0.091; interaction HR = 0.58, interaction p = 0.284. However, as the numbers in the RM+ subgroup of this ‘double wild type’ group are very small, this limits the evidence that can be drawn from these data.
The results for the EGFR wild‐type population were confirmed by looking at samples from the TITAN study of erlotinib versus chemotherapy in second‐line NSCLC 10, in which the percentage of RM FISH‐positive cases was similar, and PFS and OS were improved with erlotinib versus chemotherapy in the RM FISH‐positive group compared with the not RM FISH‐positive group (supplementary material, Figure S4; PFS: RM+ HR = 0.79, RM− HR = 1.28; OS: RM+ HR = 0.58, RM− HR = 0.92).
Ring study results
A small ring study was carried out at two reference laboratories: HistoGeneX (Antwerp, Belgium) and Targos GmbH (Kassel, Germany). Inter‐reader and between‐laboratory concordance was excellent for the overall ReadMax strategy, with a rating of 86%. Only three of the 21 samples were discordant between the laboratories, and in all three cases the more highly trained laboratory assessed the samples as positive while the laboratory that received less training scored them as negative. See supplementary material for more details.
Discussion
For a significant proportion of patients with advanced NSCLC, their tumours will not harbour either of the established predictive markers (ie EGFR mutation or EML4‐ALK fusion). There is, therefore, a considerable unmet need for a means of identifying those patients with wild‐type NSCLC who will obtain clinical benefit from specific therapies, such as EGFR TKIs. Results from trials are mixed and suggest that it is highly likely that there is a subgroup of patients who do receive benefit from erlotinib, as demonstrated in TITAN 10 and HORG 11, while other patients may respond better to chemotherapy as seen with the results from TAILOR 12 and DELTA 13, as well as the recent VeriStrat observations from PROSE 14. The identification of different patterns of EGFR gene expression could potentially allow for greater personalization of treatment choice in patients with wild‐type disease.
The first step in any FISH‐derived methodology is the setup of a reading protocol. Strategies aimed at reading a representative collection of tumour cells, like the Colorado approach, may lead to an averaging out of the actual aberrance, in particular for heterogeneous tumours. The maximization reading strategy of ReadMax strives to identify the most aberrant cells instead, and is predicated on the idea that the proved existence of aberrance within the tumour would indicate oncogene addiction.
As with all FISH testing, validation and reproducibility are key issues. However, the ReadMax system was set up after two positive feasibility studies (see supplementary material for details) and worked well in this study. Pathologists were able to extract the necessary information from the samples without difficulty.
A change in reading strategy necessitates a new scoring system, which is not automatically implied from the reading protocol. For the ReadMax scoring system, a systematic development process was implemented, considering the dangers of adaptation bias when clinical endpoints were factored in to the selection of scorings. The diagnostic development centred on the actual FISH data (without clinical endpoints), used EGFR and CEP7 counts, and used diagnostic quality assessments. Using multivariate analysis, actual patterns of EGFR aberrance in this disease population were identified. These were robust scorings, with EGFR_MULT being akin to traditional amplification and POLY_F2 describing a pattern of polysomy. Multivariate pattern recognition methods helped to identify two further polysomial patterns, based on mutation/wild‐type FISH comparison (FACS_MUT) and long PFS/early progression comparison (HI_POLY). Since the latter feature actually used clinical endpoints of a subgroup, a separate statistical adjustment reassured that no adaptation bias was discernible.
Each of the four aberrance patterns on its own showed predictive power regarding the considered endpoints, although this did not always reach statistical significance. Amplification according to EGFR_MULT was not predictive for treatment success in wild‐type patients. Although somewhat surprising, this is not a unique observation. The IPASS trial published in 2011 by Fukuoka et al. found that the value of EGFR GCN in predicting PFS benefit with gefitinib was driven by an overlap between the high EGFR GCN group and those with positive EGFR mutation status 15, which is consistent with the present finding in wild types. In future investigations, it would be interesting to investigate more closely the differential interaction of aberrance pattern with treatment success (for assistance in applying ReadMax scoring in further research, a public website is offered; see supplementary material for more details).
The ReadMax aberrance features can be combined to achieve a decision rule, which appears to work well for patients with unknown EGFR mutation status involving all four features. However, for wild types, it appeared to be more reliable to base the decision on the three polysomial factors. Indeed, when polysomial criteria were used in our analysis of SATURN data, PFS results for wild‐type patients were improved for RM FISH‐positive versus not RM FISH‐positive groups. These results were maintained when analysing the ‘double wild type’ group who were both EGFR and KRAS wild type (see supplementary material). One possibility to consider is that the four features could be differently distributed with different types of tumour histology; this is something that should be the subject of further investigation and could further refine and improve the applicability of the ReadMax scoring system.
The results seen in this study were encouraging and suggest that, with further experience, this type of testing methodology could be beneficial for clinical decision making. One of the recognized difficulties of the representative (eg Colorado) FISH scoring system (used in the original analysis of the TRUST and MERIT studies) was the challenge in inter‐laboratory diagnostic standardization. The maximization approach seems to help with this issue, as a limited ring study found that, where the ReadMax protocol was followed correctly, concordance was generally good. Even minimal training led to reproducible results, although it was clear that the training needs to emphasize the importance of the maximization reading strategy, ie scanning the whole of the sample thoroughly for the most aberrant areas. Further studies will help to confirm the applicability and reproducibility of the ReadMax strategy.
Obtaining sufficient sample tissue is a frequent issue in patients with NSCLC and testing often needs to be prioritized where limited material is available. If a reliable test was available that could use existing prepared samples, it would benefit physicians and patients alike. A limitation of the method, as with other methodologies relying on tumour samples, is that the genomic profile of a tumour can change over time. Therefore, if initial diagnostic biopsy material is used instead of more recent samples, the validity of results may be affected.
Further work on the ReadMax scoring system is required before it can be recommended for use in clinical practice. The different patterns of aberrance (in particular, the polysomial patterns) may have different impacts on erlotinib benefit, so additional research is needed to differentiate between these cellular profiles. Interestingly, although the efficacy analyses were not sufficiently powered to demonstrate statistical significance, the group of patients with RM FISH‐positive wild‐type NSCLC in SATURN (n = 36) achieved a HR of 0.63 for PFS and 0.61 for OS, in favour of erlotinib. If these results were maintained across a larger population, it would represent a clinically significant benefit, which merits further investigation. As mentioned, the four features may be differently distributed across different types of tumour histology, and understanding this could help to improve the predictive power of the ReadMax scoring system. This method also needs to be evaluated in a wider population with clear guidance and training for pathologists, in order to assess its validity and robustness.
In conclusion, the ReadMax scoring system is a promising technique, which could help to identify those patients likely to obtain clinical benefit from erlotinib treatment. If confirmed in future studies, this method could potentially optimize treatment choice, particularly for patients with EGFR wild‐type NSCLC.
Author contributions
Joachim Moecks and Barbara Klughammer contributed to the design of the study, bio‐statistical and bio‐mathematical approaches, data interpretation and analysis and writing of the manuscript. Denis Soulières discussed the analysis at an advisory meeting and contributed to the interpretation of results and writing of the manuscript.
Supporting information
Figure S1. PFS in patients with EGFR wild‐type NSCLC according to FISH status.
Figure S2. (A) PFS, (B) OS and (C) 12‐week DCR in unselected SATURN patients by ReadMax score.
Figure S3. (A) PFS, (B) OS and (C) 12‐week DCR in EGFR wild‐type SATURN patients by ReadMax score.
Figure S4. PFS and OS in the EGFR wild‐type population of TITAN according to ReadMax FISH status.
Supporting Information
Acknowledgements
Support for third‐party medical writing assistance was provided by F. Hoffmann‐La Roche Ltd.
References
- 1. Cappuzzo F, Hirsch FR, Rossi E, et al. Epidermal growth factor receptor gene and protein and gefitinib sensitivity in non‐small‐cell lung cancer. J Natl Cancer Inst 2005; 97: 643–655. [DOI] [PubMed] [Google Scholar]
- 2. Hirsch FR, Varella‐ Garcia M, McCoy J, et al. Increased epidermal growth factor receptor gene copy number detected by fluorescence in situ hybridization associates with increased sensitivity to gefitinib in patients with bronchioloalveolar carcinoma subtypes: a Southwest Oncology Group Study. J Clin Oncol 2005; 23: 6838–6845. [DOI] [PubMed] [Google Scholar]
- 3. Zhu CQ, da Cunha Santos G, Ding K, et al. National Cancer Institute of Canada Clinical Trials Group Study BR.21. Role of KRAS and EGFR as biomarkers of response to erlotinib in National Cancer Institute of Canada Clinical Trials Group Study BR.21. J Clin Oncol 2008; 26: 4268–4275. [DOI] [PubMed] [Google Scholar]
- 4. Varella‐Garcia M, Diebold J, Eberhard DA, et al. EGFR fluorescence in situ hybridisation assay: guidelines for application to non‐small‐cell lung cancer. J Clin Pathol 2009; 62: 970–977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Brugger W, Triller N, Blasinska‐Morawiec M, et al. Prospective molecular marker analyses of EGFR and KRAS from a randomized, placebo‐controlled study of erlotinib maintenance therapy in advanced non–small‐cell lung cancer. J Clin Oncol 2011; 29: 4113–4120. [DOI] [PubMed] [Google Scholar]
- 6. Soulières D, Ciuleanu T, Stelmakh L, et al. Outcomes with erlotinib in advanced NSCLC: examining the influence of increased EGFR gene copy number and EGFR mutations. Eur J Cancer 2009; 7: 556 (abstract 9167). [Google Scholar]
- 7. Tan E‐H, Ramlau R, Pluzanska A, et al. A multicentre phase II gene expression profiling study of putative relationships between tumour biomarkers and clinical response with erlotinib in non‐small‐cell lung cancer. Ann Oncol 2010; 21: 217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Reck M, van Zandwijk, N , Gridelli C, et al. Erlotinib in advanced non‐small cell lung cancer: efficacy and safety findings of the global phase IV tarceva lung cancer survival treatment study. J Thorac Oncol 2010; 5: 1616–1622. [DOI] [PubMed] [Google Scholar]
- 9. Cappuzzo F, Ciuleanu T, Stelmakh L, et al. Erlotinib as maintenance treatment in advanced non‐small‐cell lung cancer: a multicentre, randomised, placebo‐controlled phase 3 study. Lancet Oncol 2010; 11: 521–529. [DOI] [PubMed] [Google Scholar]
- 10. Ciuleanu T, Stelmakh L, Cicenas S, et al. Efficacy and safety of erlotinib versus chemotherapy in second‐line treatment of patients with advanced, non‐small‐cell lung cancer with poor prognosis (TITAN): a randomised multicentre, open‐label, phase 3 study. Lancet Oncol 2012; 13: 300–308. [DOI] [PubMed] [Google Scholar]
- 11. Karampeazis A, Voutsina A, Souglakos J, et al. Pemetrexed versus erlotinib in pretreated patients with advanced non‐small cell lung cancer: A Hellenic Oncology Research Group (HORG) randomized phase 3 study. Cancer 2013; 119: 2754–2764. [DOI] [PubMed] [Google Scholar]
- 12. Garassino MC, Martelli O, Broggini M, et al. Erlotinib versus docetaxel as second‐line treatment of patients with advanced non‐small‐cell lung cancer and wild‐type EGFR tumours (TAILOR): a randomised controlled trial. Lancet Oncol 2013; 14: 981–988. [DOI] [PubMed] [Google Scholar]
- 13. Okano Y, Ando M, Asami K, et al. Randomized phase III trial of erlotinib (E) versus docetaxel (D) as second‐ or third‐line therapy in patients with advanced non‐small cell lung cancer (NSCLC) who have wild‐type or mutant epidermal growth factor receptor (EGFR): Docetaxel and Erlotinib Lung Cancer Trial (DELTA). Proc Am Soc Clin Oncol 2013; 31: 8006. [DOI] [PubMed] [Google Scholar]
- 14. Lazzari C, Novello S, Barni S, et al. Randomized proteomic stratified phase III study of second‐line erlotinib (E) versus chemotherapy (CT) in patients with inoperable non‐small cell lung cancer (PROSE). J Clin Oncol 2013; 31: LBA8005. [Google Scholar]
- 15. Fukuoka M, Wu YL, Thongprasert S, et al. Biomarker analyses and final overall survival results from a phase III, randomized, open‐label, first‐line study of gefitinib versus carboplatin/paclitaxel in clinically selected patients with advanced non‐small‐cell lung cancer in Asia (IPASS). J Clin Oncol 2011; 29: 2866–2874. [DOI] [PubMed] [Google Scholar]
- 16. Schneider C‐P, Heigener D, Schott‐von‐Römer K, et al. Epidermal growth factor receptor‐related tumor markers and clinical outcomes with erlotinib in non‐small cell lung cancer: an analysis of patients from german centers in the TRUST study. J Thorac Oncol 2008; 3: 1446–1453. [DOI] [PubMed] [Google Scholar]
- 17. Pirker R, Su W, Rooneem, R , et al. Clinical outcomes with erlotinib in relation to biomarker status: analyses from the open‐label TRUST study in advanced non‐small‐cell lung cancer (NSCLC). Ann Oncol 2008; 19: viii100 (abstract 265P). [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. PFS in patients with EGFR wild‐type NSCLC according to FISH status.
Figure S2. (A) PFS, (B) OS and (C) 12‐week DCR in unselected SATURN patients by ReadMax score.
Figure S3. (A) PFS, (B) OS and (C) 12‐week DCR in EGFR wild‐type SATURN patients by ReadMax score.
Figure S4. PFS and OS in the EGFR wild‐type population of TITAN according to ReadMax FISH status.
Supporting Information


