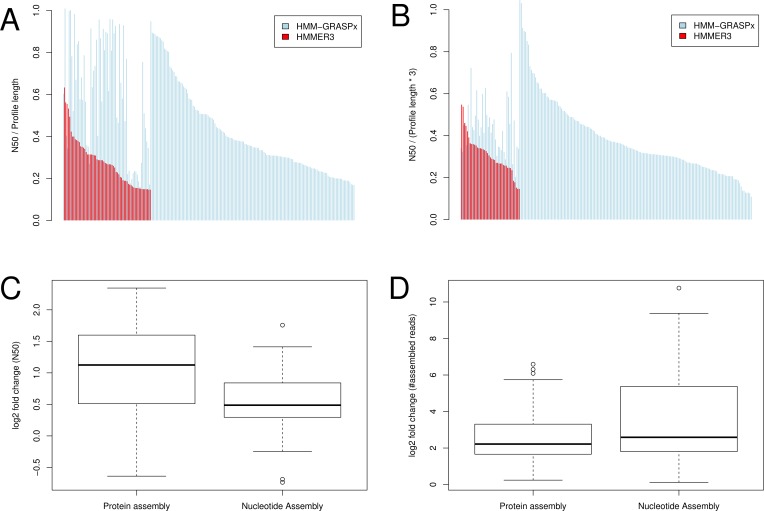
Fig 4. Targeted assembly of secondary metabolite synthesizing protein families from the in vitro human oral biofilm MT data set.
Only the protein contigs longer than 60aa and nucleotide contigs longer than 180nt were considered. (A) Normalized N50 for protein assembly. (B) Normalized N50 for nucleotide assembly. For (A) and (B), red color indicates the performance of HMMER3 and blue color indicates HMM-GRASPx. The x-axes indicate assembly cases and were sorted based on the decreasing values of the HMMER3 performance and then the decreasing values of the HMM-GRASPx performance. Assembly cases without corresponding red bars indicate that no contig was assembled using HMMER3 predictions. (C) Log2 fold change for the N50 measures in assembly cases where contigs can be assembled using either HMMER3 or HMM-GRASPx prediction. (D) Log2 fold change for the number of assembled reads in assembly cases where contigs can be assembled using either HMMER3 or HMM-GRASPx prediction.