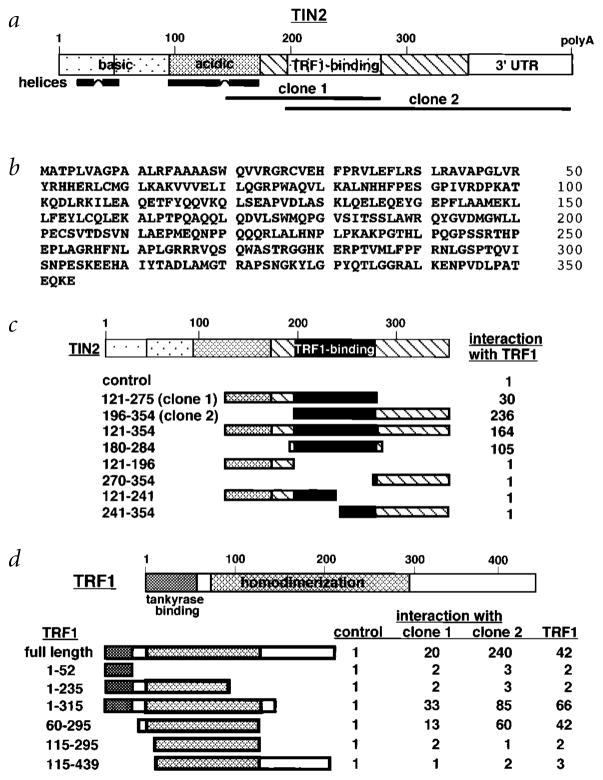
Fig. 1.
Sequence and structural characteristics of human TIN2. a, Structural features of TIN2. Shown are regions corresponding to the cDNA inserts recovered from the two-hybrid screen (clone 1, aa 147–275; clone 2, aa 196–354), the basic and acidic regions, potential helical structures and TRF1-binding domain. b, Deduced amino acid sequence of TIN2. c, TIN2 domains that interact with TRF1. We transformed TINF2 cDNA fragments (encoding the indicated amino acids) in pGAD-424 into yeast with pGBT9 containing TERF1 cDNA, and assessed interaction by a luminescent β-galactosidase assay. Control luminescence (interaction of pGAD-424 with pGBT9-TRF1) was 0.1–0.2 β-galactosidase U, and given a value of 1. We analysed 3–5 transformants for each determination. c, TRF1 domains that interact with TIN2. Depicted is TRF1, showing the tankyrase-binding and homodimerization domains. We transformed TERF1 cDNA fragments (encoding the indicated amino acids) in pGBT9 into yeast with pGAD10 containing no insert (control), TINF2 clone 1, TINF2 clone 2 or full-length TERF1 cDNA, and assessed interaction by luminescent β-galactosidase assay. Control luminescence (interaction with insertless pGAD10) was 0.1–0.2 β-galactosidase U, and given a value of 1. We analysed 3–5 transformants for each determination.