Fig. 2.
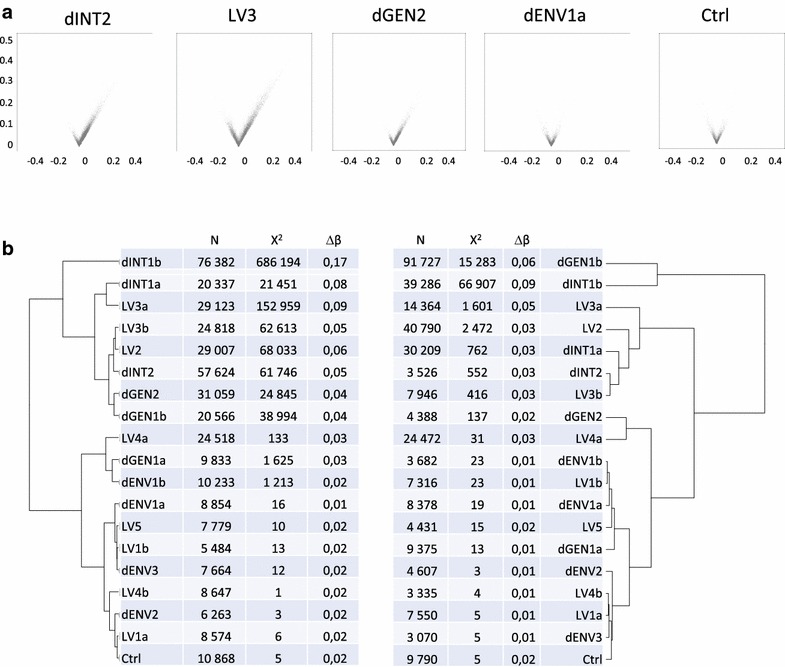
Analysis of DNA methylation changes. Analyses of 4 different chip runs (a, b, c, d) testing a total of 12 different batches of vector annotated accordingly (i.e. dENV1a and dENV1b indicates a repeat testing of dENV1 batch on chip a and chip b). a Volcano plots of representative experiments. From left to right: methylation differences observed in cells infected with an integrative lentiviral vector showing a high effect (LV2), integrase-deficient particles (dINT2), genome-deficient (dGEN2) and envelope-deficient (dENV1) particles as compared to control cells cultured but without any vector. The last plot on the right represents a control triplicate compared to another control triplicate (as in bottom line of b). Each point on a volcano plot represents the maximal delta-β value of a CpG site in a given triplicate as a function of the median of the three delta-β values of that triplicate. Only the CpG-s identified as displaying altered methylation between the samples and their corresponding controls are shown. Points representing CpG sites with increased and decreased methylation are displayed on the right and left sites of the plot. Note the higher number of points and higher median and maximal delta-β values with increased methylation in the first three samples. b Hierarchical classification of the experimental conditions depending on the extent of the increase (left) or the decrease (right) of the genomic DNA methylation in CD34+ cells. The classification was done using the three parameters indicated in the table: number of changed CpGs (N), chi2 of genomic distribution and median delta-β
