Fig. 4.
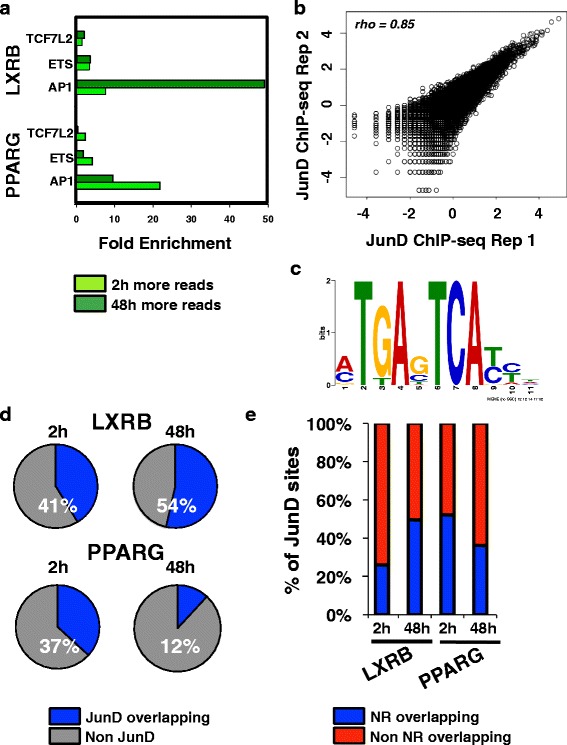
Cofactor enrichment and genome-wide binding patterns. a The fold enrichment of various canonical motifs for the top 50 % of significance-ranked LXRB and PPARG binding sites that exhibited stronger occupancy at 2 (light green) or 48 (dark green) h. b Normalized sequencing read depth rank correlation (top left) at all binding sites identified across replicate JunD ChIP-seq experiments. c Enriched AP1 motif identified at JunD ChIP-seq binding sites. d Percentage of LXRB and PPARG sites after 2 and 48 h of stimulation coincident with JunD. e Percentage of JunD sites overlapping with LXRB and PPARG nuclear receptor (NR) binding events after 2 and 48 h of drug treatment
