Figure 5. Predicted locations of LILRB3 and LILRA6 polymorphisms that alter binding to necrotic glandular epithelial cells, and sites under strong evolutionary selection pressure.
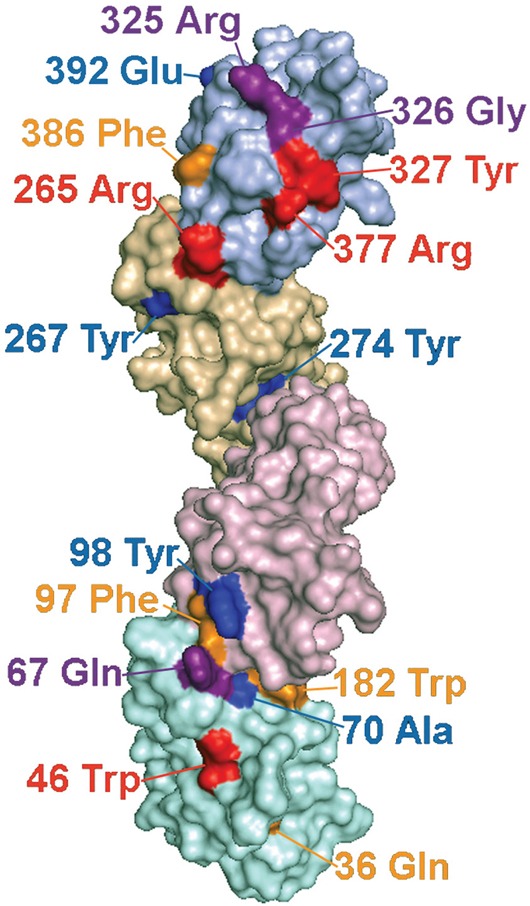
Molecular model of LILRB3 based on the structure of related LILR proteins. Ig domains are shaded in different colours with D1 at the bottom of the figure and D4 at the top. The labelled polymorphic residues are coloured to indicate their influence on ligand binding and selection pressure, as follows: Red= Polymorphic amino acids implicated in ligand binding that have undergone significant positive selection pressure; Purple= Putative ligand binding amino acids that are not under significant positive selection; Yellow=Amino acids that are undergoing positive selection, but do not appear to influence ligand binding; Blue=Amino acids undergoing negative selection. Amino acid 326 (coloured purple) is under significant negative selection and also implicated in binding.
