Figure 2. Summary of compounds whose transcriptional response is similar to that induced by the knock-down of the TNF Network.
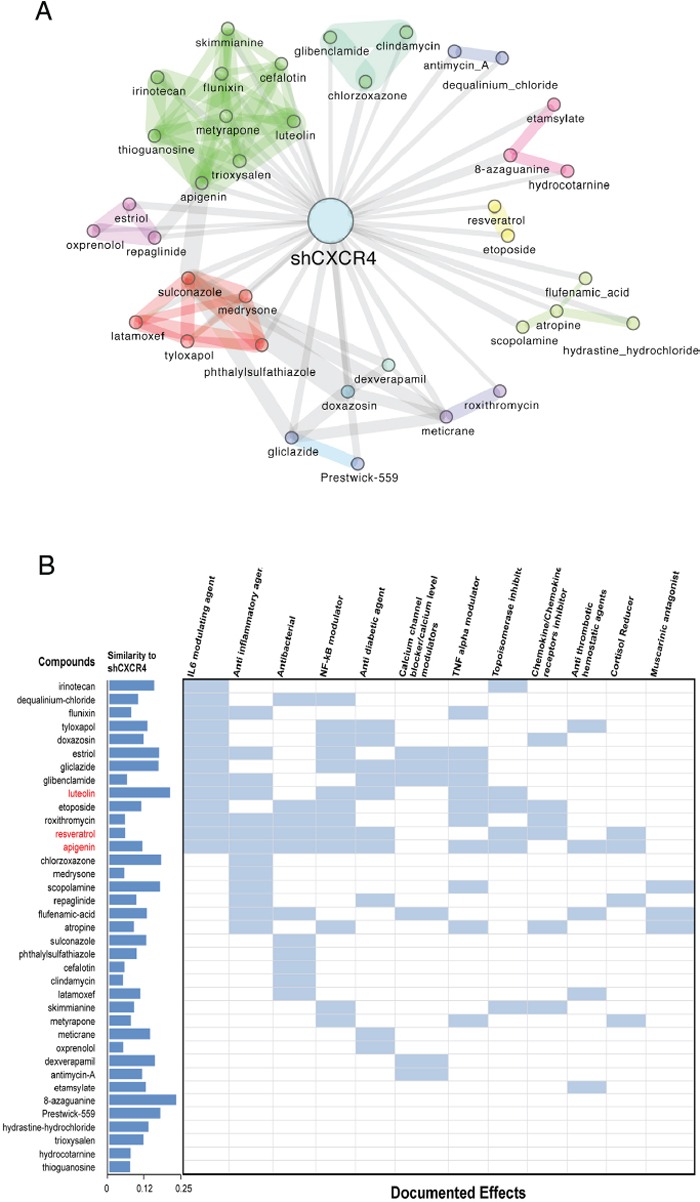
A. Drug-network based classification using the gene expression signature in shCXCR4 cells. The expression signature of cells in which the TNF network has been inhibited is connected to a given drug if the profile of differential expression following the knock-down is significantly similar to the transcriptional response elicited by that drug in human cancer cell lines. Two drugs are connected to each other if their transcriptional response is similar according to the same criteria. Colors indicate groups of densely interconnected nodes enriched for a given mode of action, common drug targets or therapeutic application. B. Included are documented effects for each drug in light blue and shown in red are the drugs known to target protein kinase CK2. The dark blue bars indicate the extend of similarity between the transcriptional responses elicited by the listed compounds and that of the knock-down of CXCR4 (shCXCR4). This is defined as the inverse of the distance reported in Supplementary Table 1, and normalised in [0,1]. According to this metric the compound eliciting the most similar response to shCXCR4 is 8-azaguanine and CK2 is a recurrent target among those of the top-similar to shCXCR4.
