Figure 4. Functional genetic screen with AMPI-109 identifies PRL-3 amplification in invasive basal breast cancers.
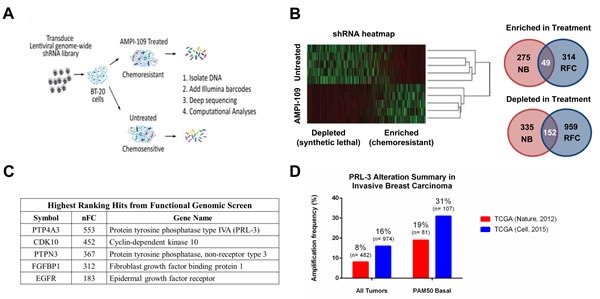
A. Schematic representation of the experimental work flow of the functional genomic screen. BT-20 cells were transduced with a genome-wide shRNA library and treated with AMPI-109 (750 nM) or left untreated. After 5 passages the shRNA tag sequences were recovered and quantified by deep sequencing. Data were analyzed by complementary computational analyses generating lists of mediators of chemoresistance and chemosensitivity to AMPI-109. B. Heatmap of shRNA tag representation in AMPI-109-treated and untreated samples. Right panel: Two analysis strategies (NB, negative binomial and RFC, relative fold change) implicated 2,084 genes that modulate the effects of AMPI-109. 201 genes were identified by combined analysis strategies. C. Table highlighting highest ranking gene hits enriched by AMPI-109 treatment by normalized fold change (nFC). D. Bar graph depicting level of PRL-3 amplification in TCGA Nature, 2012 cohort (red) and TCGA Cell, 2015 dataset (blue) for patients with invasive breast cancer (all tumors) and those with invasive basal breast cancer as determined by the PAM50 signature (PAM50 Basal).
