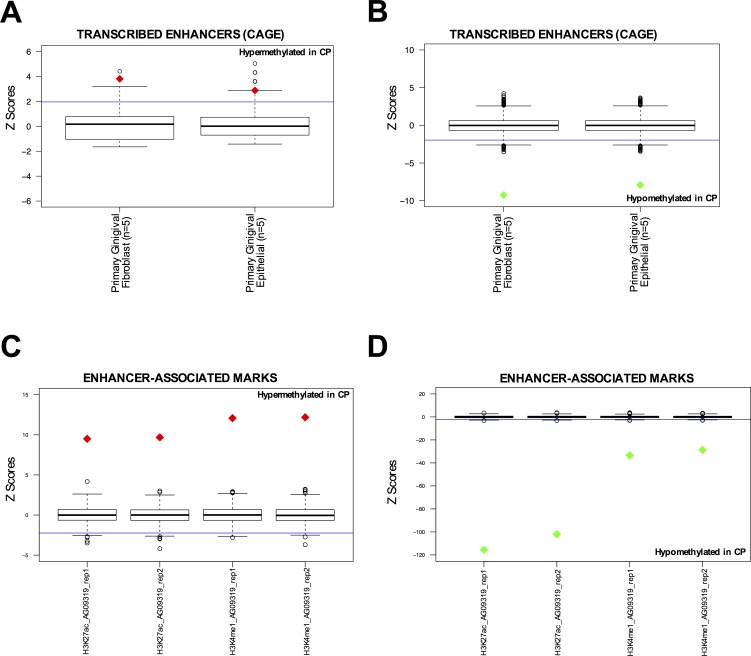
Figure 2. DNA hypermethylated sites in chronic periodontitis are enriched at normal enhancer elements in gingival tissues.
A.-B. Overlap between differentially methylated CpG sites and active enhancers (defined by bi-directonal CAGE-seq profile [28]) in primary gingival fibroblasts (n = 5) and primary epithelial (n = 5) tissues from FANTOM5 project. Overlap was counted between each differentially methylated CpG site and the called enhancer by FANTOM5 [28]. Box-plots represent 1,000 random permutations across the array of the same number of hypermethylated probes (left) or hypomethylated probes (right). Red diamonds represent the Z-score of significantly enriched marks and green diamonds represent Z-scores of significantly depleted marks. C.-D. Overlap between differentially methylated CpG sites and enhancer-associated marks (H3K27ac and H3K4me1) in normal gingival fibroblast (AG09319). ChIP-seqs were performed in-house, in duplicate. Overlap was counted between each differentially methylated CpG site and the peak for each mark called using MACS. Box-plots represent 1,000 random permutations across the array of the same number of hypermethylated probes (left) or hypomethylated probes (right). Red diamonds represent the Z-score of significantly enriched marks and green diamonds represent Z-scores of significantly depleted marks.