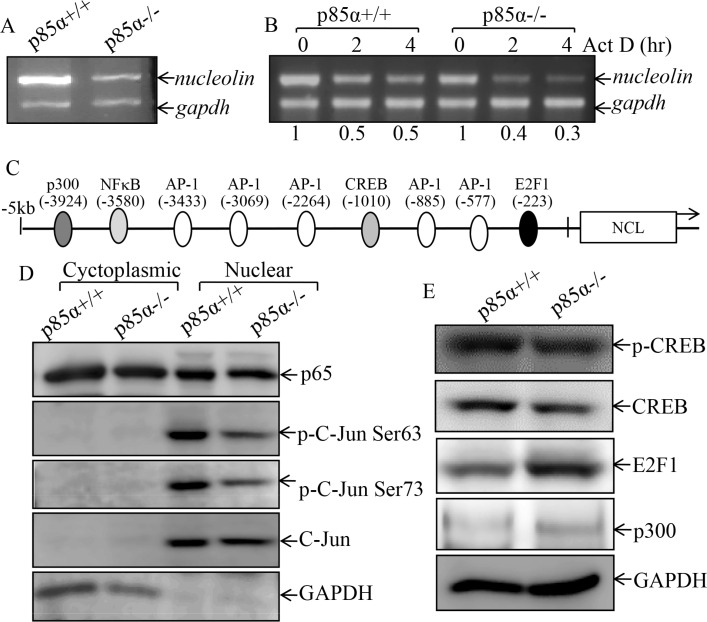
Figure 5. p85α regulated NCL transcription and C-Jun activation.
(A) p85α+/+ and p85α−/− cells were cultured in 6-well plates till cell density reaching 80–90%, and then extracted for total RNA with Trizol reagent. RT-PCR was used to determine nucleolin mRNA expression, while gadph was used as an internal control, (B) p85α+/+ and p85α−/− cells were cultured in 6-well plates till cell density reaching 80–90%, and the cells were then treated with Act D for the indicated time points, and were then used for total RNA isolation. The total RNA was subjected to RT-PCR for determination of mRNA levels of Egfr and Gapdh. (C) Potential transcriptional factor binding sites in NCL promoter region (−5000-+1) analyzed by using the TRANSFAC 8.3 engine online. (D) p85α+/+ and p85α−/− cells were cultured in 6-well plates till cell density reaching 80–90%, the cells were extracted and the whole cell extracts were used to isolate cytoplasmic and nuclear fractions according to the protocol of the nuclear/cytosol fractionation kit. The isolated protein fractions were subjected to Western blot. GAPDH were used as control for protein loading. (E) p85α+/+ and p85α−/− cells were cultured in 6-well plates till cell density reaching 80–90%, the cells were extracted and cell extracts were subjected to Western blot for determination of the indicated protein expression with specific antibodies and GAPDH was used as protein loading controls.