Fig. 2.
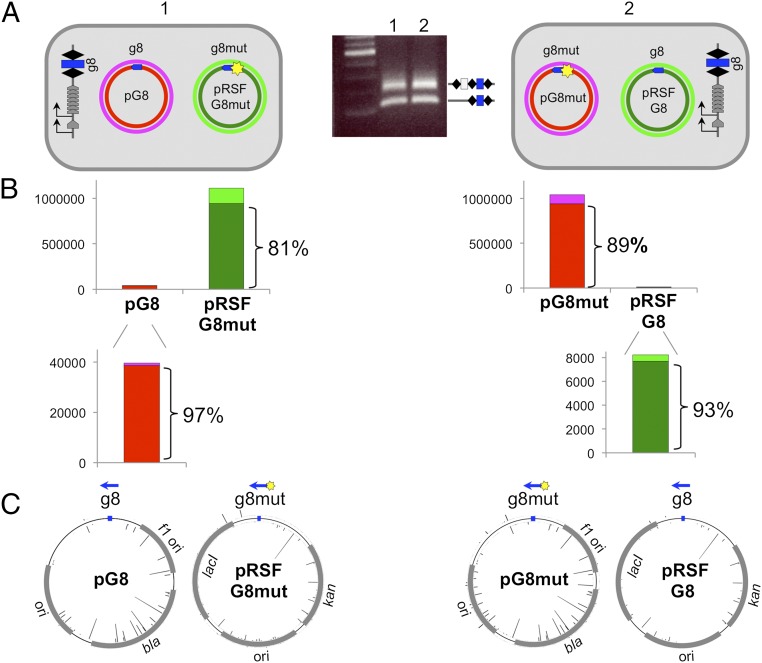
CRISPR adaptation from matching and partially matching protospacer targets located in an in trans configuration. (A) The E. coli KD263 cells transformed with compatible pT7Blue or pRSF-based plasmids each carrying a wild-type or a mutant, C1T version of the g8 protospacer are schematically shown. Protospacers are indicated as blue arrows located on the nontargeted DNA strand of each plasmid, with the arrow pointing away from the ATG PAM. Results of a CRISPR adaptation experiment with the cells transformed with two plasmids (culture 1 and culture 2) are shown in the middle. (B) Results of overall mapping of spacers acquired by cells shown in A to separate strands of each plasmid are shown. Numbers on vertical axes show the number or reads. The color code used shows the origin of spacers and matches colors used to indicate the individual plasmid strands in A. (C) Mapping of individual acquired spacers on donor plasmids. Blue arrows indicate the position of the g8 protospacer. Bars protruding inside and outside the circles indicating plasmids represent spacers derived from different strands. Bar height indicates the relative efficiency of spacer acquisition from this position.