Fig. 3.
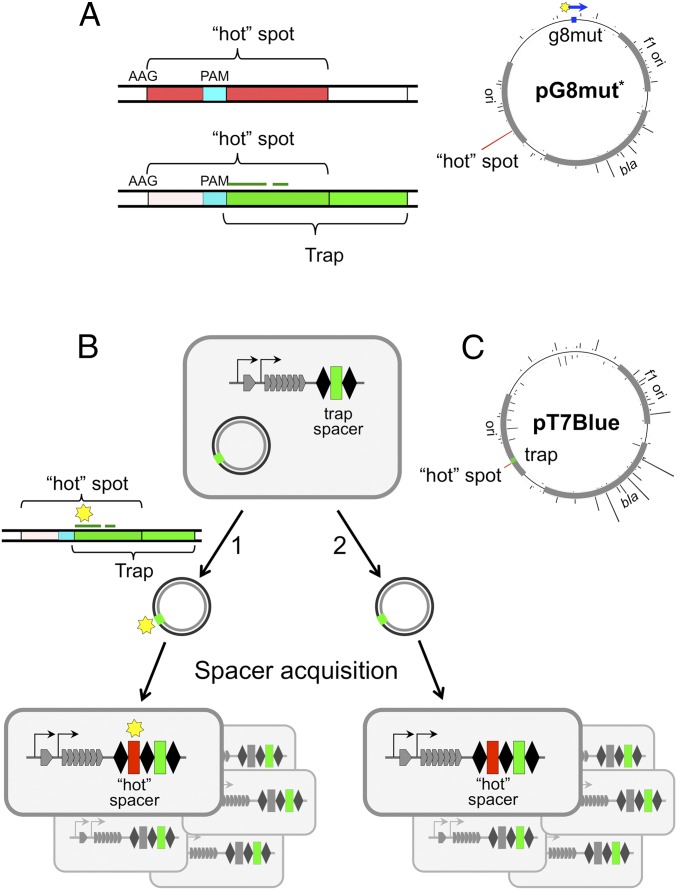
Using a trap spacer to show adaptation from fully matching protospacers. (A) Mapping of spacers acquired by KD263 cells transformed with pG8mut* are shown [note that the orientation of the protospacer in pG8mut (Figs 1. and 2) and in pG8mut* is different]. The height of bars emanating from a circle representing pG8mut* shows the relative amounts of spacers acquired from this site. The sequence of a hot protospacer (indicated by a red bar) is expanded. It contains an internal PAM sequence (cyan) defining a trap protospacer shown in green. The position of the trap protospacer seed with respect to the partially overlapping hot protospacer is shown by a thin green line. (B) The E. coli KD384 cells containing a trap spacer in their CRISPR array and transformed with the pT7Blue plasmid are schematically shown to the left. At the right, structures of CRISPR arrays of cells that acquired a hot spacer (red) from a protospacer overlapping with protospacer recognized by trap crRNA are schematically shown. (C) Mapping of spacers acquired by KD384 cells transformed with pT7Blue. The position of the priming trap protospacer is shown as a green rectangle. Red bar shows observed efficiency of spacer acquisition from the overlapping hot protospacer.