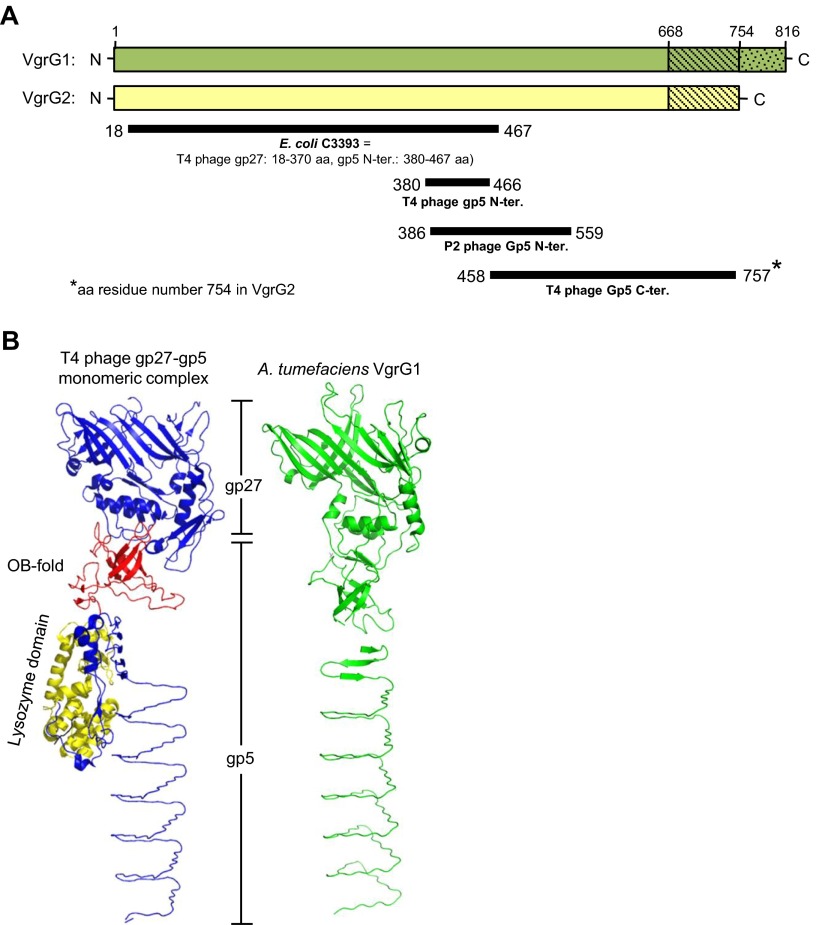
Fig. S2.
Domain prediction and structure modeling of VgrG1 and VgrG2. (A) Graphic representation of the domains predicted to present in VgrG1 and VgrG2 by the Phyre2 web server. VgrG1 and VgrG2 full-length proteins are represented as green and yellow bars, respectively (drawn to scale). Solid dark lines represent the region of VgrG1/VgrG2 that is structurally similar (confidence > 99%) to the protein shown in bold under each line. The number at the start and end of each line indicates the amino acid position from VgrG1/VgrG2. According to ref. 14, E. coli C3393 protein is structurally similar to the gp27 protein and the N-terminal of gp5 protein in T4 phage. (B) Structure comparison of A. tumefaciens VgrG1 and phage gp27-gp5 monomeric complex derived from the gp5–gp27 trimeric complex (PDB ID code 1K28). Shown for VgrG1 the best-predicted model among nine different models generated using PHYRE2, SWISS-MODEL, and I-TAESSER homology modeling servers. In the gp27–gp5 monomeric complex structure, the gp27 domain and gp5 C terminus is shown in blue, the OB fold in red and the lysozyme domain in yellow. The predicted structure of A. tumefaciens VgrG1 is in green.