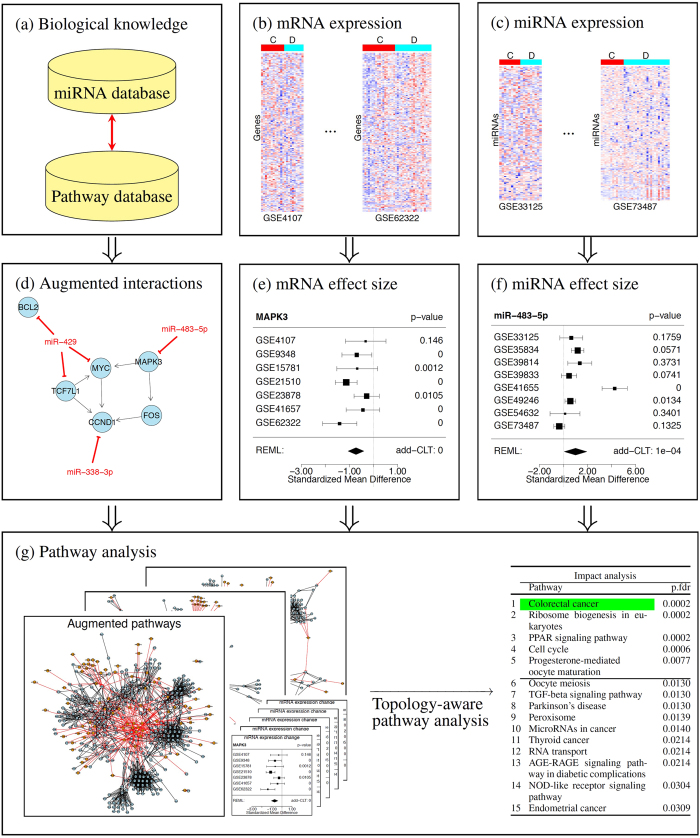
Figure 1. Overall pipeline of the proposed framework.
The input consists of (i) a pathway database and a miRNA database including known targets (panel a), (ii) multiple mRNA expression datasets (panel b), and (iii) multiple miRNA expression datasets (panel c). Each expression dataset consists of two groups of samples, e.g. disease versus control. The framework first augments the signaling pathways with miRNA molecules and their interactions with coding mRNA genes (panel d). It then calculates the standardized mean difference and its standard error in each expression dataset. The summary size effect across multiple datasets for each data type are then estimated using the REstricted Maximum Likelihood (REML) algorithm (panels e,f). Similarly, the p-value for differential expression is calculated for each dataset and then combined using the additive method (add-CLT). The augmented pathways, the combined p-values, and the estimated size effects then serve as input for ImpactAnalysis, which is a topology-aware pathway analysis method (panel g).