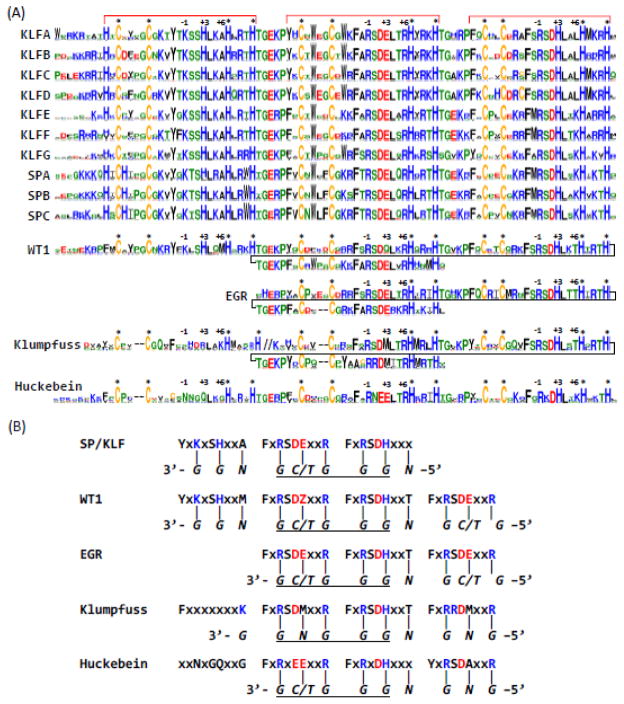
Figure 1. Sequence logos and DNA-binding preferences of Znfs of SP/KLF, WT1, EGR, Huckebein, and Klumpfuss.
(A) Sequence logos of SP/KLF groups, WT1, EGR, Huckebein and Klumpfuss. Znf regions are marked by red brackets on top. Up to eight positions before the first Znf were also shown as they provide some discrimination power among KLF and SP groups. Three key DNA-binding specificity-determining positions are marked by −1, +3 and +6 under the brackets. Zinc-binding positions with conserved cysteines and histidines are marked by stars (*). The last Znf of WT1, EGR, and Klumpfuss is aligned to the second Znf of SP/KLF and WT1. The coloring of amino acids is as follows: C – yellow; R,K,H – blue; D,E – red; W,F,Y,M,L,I,V,A – black; G,P,S,T, N, Q – green. The linker region between the first Znf and the second Znf of Klumpfuss is replaced with “//”, as it does not conform to the TGE[RK]P consensus in terms of length and amino acid composition. These sequence logos were obtained by using the WebLogo 3 server (Crooks et al. 2004). (B) Predicted DNA-binding preferences of SP/KLF, WT1, EGR, Huckebein and Klumpfuss. The motifs harboring key DNA recognition positions are shown for Znfs of each family. The preferred DNA sequences are shown in italic letters under the motifs with the direction marked by 5′ and 3′ at the ends. Each base was under its corresponding interaction position in the protein motif. The core GC/GT-box sequences are underlined. The last Znf of Huckebein is in grey letters as it is missing in many Huckebein proteins. The letter Z in the second Znf motif of WT1 denotes E or Q. Metazoan and nonmetazoan WT1s have Q and E in this position, respectively.