Abstract
Background The threat posed by swine influenza viruses with potential to transmit from pig populations to other hosts, including humans, requires the development of new experimental systems to study different aspects of influenza infection. Ex vivo organ culture (EVOC) systems have been successfully used in the study of both human and animal respiratory pathogens.
Objectives We aimed to develop an air interface EVOC using pig tracheas in the study of influenza infection demonstrating that tracheal explants can be effectively maintained in organ culture and support productive influenza infection.
Methods Tracheal explants were maintained in the air interface EVOC system for 7 days. Histological characteristics were analysed with different staining protocols and co‐ordinated ciliary movement on the epithelial surface was evaluated through a bead clearance assay. Explants were infected with a swine H1N1 influenza virus. Influenza infection of epithelial cells was confirmed by immunohistochemistry and viral replication was quantified by plaque assays and real‐time RT‐PCR.
Results Histological analysis and bead clearance assay showed that the tissue architecture of the explants was maintained for up to 7 days, while ciliary movement exhibited a gradual decrease after 4 days. Challenge with swine H1N1 influenza virus showed that the EVOC tracheal system shows histological changes consistent with in vivo influenza infection and supported productive viral replication over multiple cycles of infection.
Conclusion The air interface EVOC system using pig trachea described here constitutes a useful biological tool with a wide range of applications in the study of influenza infection.
Keywords: Air interface, ex vivo organ culture, influenza, swine
Introduction
Influenza A virus is the most significant worldwide respiratory pathogen, infecting a range of species that includes humans, pigs, horses, sea mammals and birds. 1 Influenza viruses are divided into different subtypes based on the antigenic reactivity of their two surface glycoproteins, the haemagglutinin and the neuraminidase. So far, 16 different haemagglutinin (H1–H16) and nine neuraminidase (N1–N9) subtypes have been identified among all influenza A viruses. 2 Waterfowl, shorebirds and gulls are considered the natural hosts of all influenza subtypes, from which mammalian influenza viruses are directly or indirectly derived. 1 , 3
Influenza infection in humans and pigs is primarily restricted to the upper and lower respiratory tract with viral replication occurring in the epithelial cells present on the surface of the respiratory mucosa. 4 The acute phase of infection is characterized by a multifocal destruction of the epithelium with cellular desquamation to the luminal space, which may often lead to patches along the mucosa with few or no epithelial cells over the basement membrane. It is also associated with the presence of oedema, vascular congestion, hyperaemia and infiltration of inflammatory cells into the submucosa underlying the epithelial layer. 4 , 5 The presence of both N‐acetylneuraminic acid‐α2,3‐galactose (NeuAcα2,3Gal) and NeuAcα2,6Gal receptors in the pig respiratory tract makes this species susceptible to most human and avian influenza viruses. 6 , 7 For this reason, pigs have been proposed as a ‘mixing‐vessel’ for reassortment between human and avian viruses. 3 , 6 , 7 , 8 The significance of cross‐species transmission of swine influenza viruses to humans is underscored by the recent human influenza outbreak with an H1N1 of possible swine origin. 9 , 10
In addition to being important natural hosts, pigs constitute a valuable animal model for the study of influenza infection, 5 , 11 as virus may be transmitted experimentally to in‐contact animals and they can exhibit clinical signs, histopathology and cytokine expression profiles comparable to influenza infection in other species. 3 , 4 , 5 , 11 , 12 In vivo studies have provided valuable information on clinical course, viral pathogenesis, innate and acquired immune responses, vaccine efficacy and antiviral treatment. 4 , 5 , 13 , 14 , 15 However, such studies are greatly affected by animal welfare considerations and logistical constraints. To overcome such constrains, in vitro systems that involve either established or primary cell lines have been widely used, 16 , 17 but such systems usually lack the natural physiological context present in the respiratory mucosa, such as cell polarization, coordinated ciliary activity, mucus production and apical contact with air.
Ex vivo organ cultures (EVOC) of tracheal explants with an air interface system have been successfully developed and used in the study of both human and animal respiratory pathogens. 18 , 19 , 20 , 21 , 22 , 23 , 24 , 25 , 26 , 27 , 28 , 29 , 30 Such systems reproduce, to a great extent, the physiological conditions and the cellular complexity of the respiratory mucosa of the host in a highly controlled setting. The three‐dimensional structure and cell diversity of the respiratory mucosa is preserved together with important physiological features like normal expression levels of cell receptors, effective mucus production and ciliary activity. 18 , 23 , 27 Moreover, EVOC systems allow a quantitative analysis of the effects of pharmacological agents (i.e. antiviral drugs) and/or the host innate response on infection dynamics. 23 , 27 , 31 In addition, tissue collected from a single animal can be used in numerous replicas, which reduces the experimental variability and the number of animals used, following the principles of (reduction, replacement and refinement (3Rs) in animal experimentation. 32
In this study, we developed a swine tracheal EVOC system for the study of influenza virus infection. Swine tracheal explants infected with a contemporary avian‐like swine H1N1 virus supported productive infection and exhibited histopathological changes consistent with in vivo influenza infection. Overall, this system constitutes a useful physiologically relevant model for the study of influenza infection in a scalable, well‐defined and controlled experimental setting.
Material and methods
Air interface EVOC of pig trachea
Tissue collection and preparation
Weaned pigs (6–8 weeks old), sourced from a Specific Pathogen Free (SPF) closed herd serologically negative against influenza virus, were killed by intravenous administration of sodium pentobarbitone (0·8 mg/kg i.v. to effect). Whole tracheas were aseptically collected and transported in pre‐warmed culture medium consisting of a 1:1 mixture of Dulbecco’s modified Eagle’s medium (DMEM) and Roswell Park Memorial Institute (RPMI) 1640 medium supplemented with penicillin (100 U/ml; Sigma), streptomycin (50 μg/ml; Sigma, Dorset, UK), l‐glutamine (10 mm; Sigma) and amphotericin (2·5 μg/ml; Sigma). Once in the laboratory, tracheas were kept at 37°C in a 5% CO2–95% air mixture in a humidified incubator. Four to six washes were performed, over a period of 3 hours, by immersing the tissue in fresh, warm DMEM/RPMI medium. The presence of bacterial flora was evaluated by inoculation of the final washing medium in Brain Heart Infusion Broth (Oxoid, Hampshire, UK) with further incubation at 37°C for 72 hours.
Preparation of the tracheal explants
After the washing steps, the surrounding connective tissue present exterior to the tracheal cartilage was removed and tracheas were opened and cut lengthways into four strips. Each segment, incorporating the respiratory mucosa and underlying cartilage, was then cut into approximately 1·0 × 0·5 cm explants and placed on a square section of sterile filter paper, which was in turn placed onto agarose plugs in 6‐well plates with the medium bathing the filter paper and the basal portion of the explants, as described previously. 29 Explants were maintained at 37°C in a 5% CO2–95% air mixture in a humidified incubator for up to 7 days.
Histological evaluation
Tissue samples were fixed in 10% (v/v) buffered formalin and paraffin‐embedded. Five micrometre sections from across each explant were subject to haematoxylin–eosin, periodic acid‐Schiff and Van Gieson staining.
Epithelial layer thickness
Haematoxylin–eosin stained slides were used to measure the thickness of the epithelial layer using ImageJ software (http://rsbweb.nih.gov/ij/). Five randomly selected fields across each section were measured and statistical significance was assessed by analysis of variance using Prism 5 (Graphpad Software, La Jolla, CA, USA).
Bead clearance assay
Five microlitres of an emulsion of 1 μm diameter polystyrene microsphere beads (Polybead microspheres, Polysciences Inc., Northampton, UK) were pipetted onto the epithelial surface of the explants, and bead clearance was evaluated at 10, 20 and 30 minutes post‐inoculation. The assay was considered positive if the microbeads had been completely cleared to one edge of the tissue.
Organ culture infection
Virus
A stock of avian‐like swine H1N1 influenza virus (A/swine/England/453/06) was grown in embryonated chicken eggs, aliquoted and stored at −80°C. The virus was isolated at the Veterinary Laboratories Agency (VLA) from pigs suffering a respiratory disease outbreak in England in 2006. Viral titres were determined by haemagglutination assays using adult chicken erythrocytes and plaque assays on Madin‐Darby Canine Kidney (MDCK) cells. 33
Organ culture inoculation
Viral inoculations were performed in triplicate in three independent experiments, with doses ranging from 2·5 × 101 to 2·5 × 106 plaque forming units (pfu). Five microlitres of viral suspension was applied onto the epithelial surface of the explants. Culture medium was used for mock‐infected controls. Inoculated explants were sampled every 24 hours for histology, bead clearance assays and viral quantification.
Immunohistochemistry
Re‐hydrated sections were treated with 3% H2O2 in methanol for 15 minutes at 22 ± 3°C for endogenous peroxidase blocking, washed twice in tap water and incubated with Pronase XXIV (MenaPath, Wokingham, UK) for 10 minutes at 22 ± 3°C for antigen retrieval. After two washes with distilled water, sections were blocked with 1·5% donkey serum in Tris Base Saline buffer (TBS) for 20 minutes and incubated overnight at 4°C with a chicken anti‐swine influenza polyclonal antibody (1:4000) kindly provided by VLA‐Weybridge. Slides were washed twice for 5 minutes with 0·05% Tween 20 in TBS (TBS/T) and incubated for 1 hour at 22 ± 3°C with a donkey anti‐chicken IgY biotin‐labelled antiserum (1:200 in blocking buffer; Jackson Immunoresearch Europe Ltd, Newmarket, UK). Sections were washed twice in TBS/T, incubated for 30 minutes with Vectastain Elite ABC reagent (Vectastain Elite ABC Kit, Vector Laboratories Peterborough, UK) prepared following the manufacturer’s instructions, then washed twice with TBS/T (5 minutes each). Colour development was carried out by incubation with SIGMAFAST™ solution (Sigma) for 10 minutes. Slides were counterstained with Mayer’s haematoxylin, dehydrated and mounted with DPX mounting medium.
Virus isolation
Inoculated explants were immersed in 1·5 ml of sterile phosphate‐buffered saline (pH 7·4) and shaken in a Mixer Mill MM300 (Retsch, Leeds, UK) at 20 Hz for 10 minutes at 4°C, followed by centrifugation at 16 200 g for 10 minutes. The collected supernatant was used in plaque assays (performed in triplicate) in MDCK cells. Results were expressed as arithmetic mean pfu/ml.
Quantification of viral copy number by real‐time RT‐PCR
RNA extraction and reverse transcription. Inoculated explants were disrupted in 1 ml of Trizol® Reagent (Invitrogen) using a Mixer Mill® MM300 (Retsch, Leeds, UK). RNA extractions were performed according to the manufacturer’s instructions, with slight modifications: after the phase separation step, the aqueous phase containing the RNA fraction was transferred to a new tube and purified with RNeasy Mini Kit (Qiagen, Crawley, UK). cDNA synthesis from 50 ng of total RNA was performed with SuperScript™ III Reverse Transcriptase (Invitrogen, Paisley, UK), according to the manufacturer’s instructions, using the specific primer for the influenza matrix segment described in Hoffmann et al. 34
Real‐time RT‐PCR. Viral copy numbers were estimated using a real‐time quantitative RT‐PCR (qPCR) assay 35 run in a Corbett Rotor Gene 3000 (Corbett Research, Mortlake, Australia). Cycle parameters were: 15 minutes at 95°C, followed by 45 cycles of 15 seconds at 94°C and 60 seconds at 60°C, with fluorescence acquisition in the last step. Standard curves were generated using 10‐fold dilutions of a plasmid containing the target sequence of the influenza matrix segment, ranging from 1 × 102 to 1 × 108 copies per microlitre. For each run, all samples, as well as the no‐template controls, plasmid standards, and positive and negative controls were run in triplicate and expressed as the arithmetic mean number of vRNA copies per microlitre of cDNA.
Results
Tracheal explants maintain normal histological architecture for up to 7 days, with progressive loss of ciliary activity
Uninfected tracheal explants preserved their normal histology without major alterations for up to 7 days after removal from the animal (Figure 1A–E), with signs of sporadic cell degeneration in the transitional layers between the epithelium and the submucosa. The overall structure of both the pseudostratified columnar epithelium, and the underlying lamina propria were maintained. A progressive increase in the amount of mucus on the epithelial apical surface was observed from day 3 onwards (Figure 1D,E). Periodic acid‐Schiff staining showed that explants maintained overall carbohydrate levels (Figure 2A–D). Van Gieson staining showed that the elastic fibres present in the submucosa had a wave‐like form after day 4 indicating a small level of fibre relaxation (Figure 2E–H). The average thickness of the epithelium did not change significantly (P = 0·4) for the duration of the organ culture (Figure 3A). A bead clearance assay was used to evaluate co‐ordinated ciliary activity. Tracheal explants were tested immediately after collection and every 24 hours thereafter. Beads were cleared in less than 30 minutes in all pieces examined. However, a progressive increase in the time for complete bead clearance was observed from day 4 onwards (Figure 3B).
Figure 1.
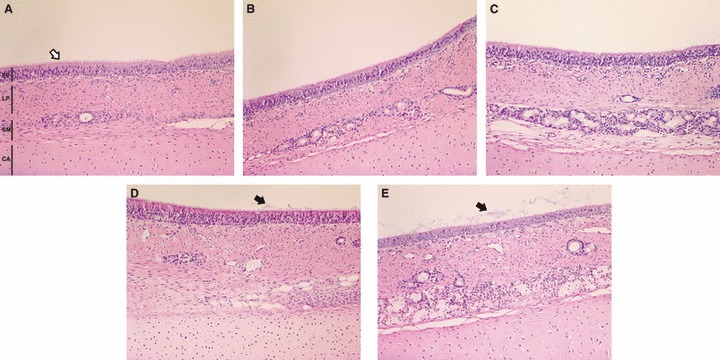
Representative light photomicrograph of sections of explants maintained in the ex vivo organ culture system for 0 (A), 1 (B), 3 (C), 5 (D) and 7 days (E). EP ‐ epithelium; LP ‐ lamina propria; SM ‐ submucosa; CA ‐cartilage. Sections were stained with haematoxylin and eosin (200× magnification). Cilia (white arrow) and mucus (black arrow) are shown in the apical surface of epithelial cells.
Figure 2.
Representative light photomicrograph of sections of explants maintained in the ex vivo organ culture system for 0 (A, E), 1 (B, F), 4 (C, G) and 7 days (D, H). Sections were stained with periodic acid‐Schiff (A–D), black arrows indicating goblet cells, or Van Gieson (E–H) showing elastic fibres stained black in the submucosa (200× magnification).
Figure 3.
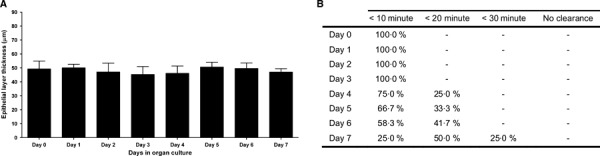
Mean epithelial layer thickness (A) and percentage of tracheal explants clearing microbeads within specified times (B) over the course of 7 days in the ex vivo organ culture system. Data are represented as means + SD (error bars) of three independent experiments.
Infected swine tracheal explants display similar histopathology to that observed in in vivo influenza infection
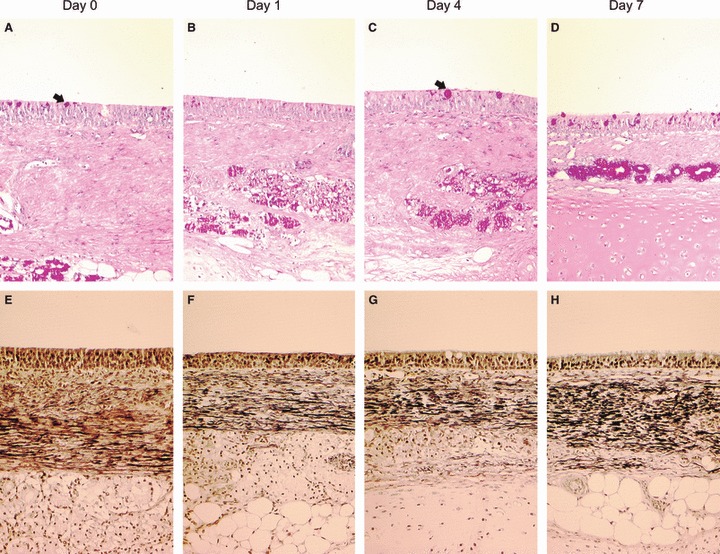
Tracheal explants were infected with avian‐like swine H1N1 influenza virus (A/swine/England/453/06). A high dose (2·5 × 106 pfu) was initially used to attempt to ensure viral infection of the majority of susceptible cells. Mock‐infected pieces displayed normal histological appearance as described above (Figure 4A,C). Infected explants (Figure 4B,D) exhibited histopathological changes compatible with in vivo influenza infection, 4 , 36 including loss of cilia at the apical surface, shrinking and vacuolization of epithelial cells with signs of destruction and desquamation and the presence of cellular debris in the mucus of the epithelial surface. Furthermore, a significant difference in the thickness of the epithelial layer between mock and virus‐infected was observed (P < 0·0001) with a marked reduction at day 1 post‐infection (pi) (Figure 4E). Ciliary‐mediated bead clearance was reduced 24 hours pi and completely absent at day 5 (Figure 4F). Viral antigen was detected in infected explants by immunohistochemistry. The distribution of positive cells was relatively widespread throughout the whole section and was limited to the epithelial layer (Figure 5A). Positive staining was predominantly seen in the nucleus, but with some also in the cytoplasm (Figure 5B). Viral antigen was also detected between the cilia and in aggregates of mucosal material present at the surface of the epithelium. In subsequent infections with a lower dose of virus (2·5 × 102 pfu) the pattern of cells with positive staining remained evenly distributed across the tissue section (Figure 5C,D).
Figure 4.
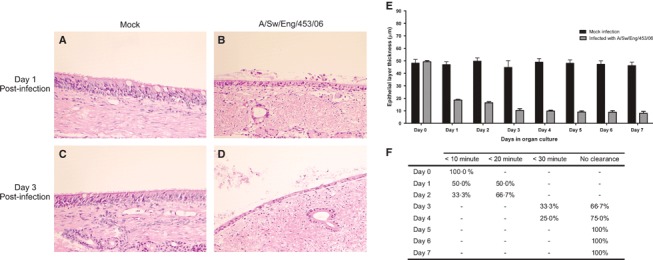
Representative light photomicrographs of sections from swine trachea stained with haematoxylin–eosin (A–D). Mock infected explants at day 1 (A) and day 3 (C) post‐infection compared with influenza‐infected explants (B, D) showing loss of cilia at the apical surface with signs of destruction and desquamation of epithelial cells and the presence of cellular debris in the mucus of the epithelial surface (200× magnification). Mean epithelial thickness measurements of mock‐infected and infected explants (E) and percentage of infected tracheal explants clearing microbeads within specified times (F) over the course of 7 days. Data are represented as mean + SD (error bars) of three independent experiments.
Figure 5.
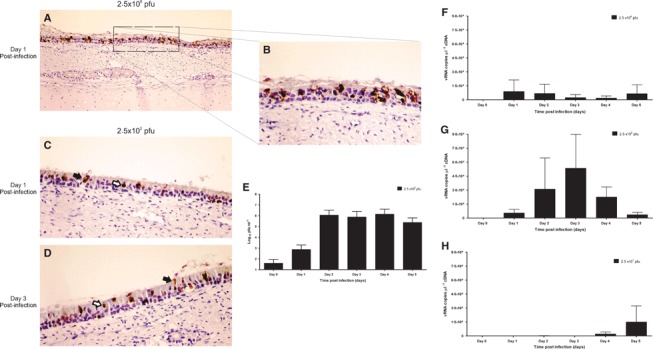
Representative light photomicrograph of sections of explants infected with H1N1 swine influenza virus stained by immunohistochemistry and counterstained with Mayer’s haematoxylin. Day 1 post‐infection (pi) with 2·5 × 106 pfu (A), with a close‐up (B). Day 1 (C) and day 3 (D) pi with 2·5 × 102 pfu. Swine influenza antigen is stained brown. [200× (A) and 400× (B–D) magnification]. White arrows show nuclear staining and black arrows cytoplasmic antigen staining. Viral quantification of extracellular infectious virus by plaque assay in MDCK cells (E) with 1·25 × 102 pfu inoculum and by quantitative real‐time PCR with an inoculum of 2·5 × 104 (F), 2·5 × 102 (G) and 2·5 × 101 pfu (H). Day 0 corresponds to a time point immediately after inoculation. Data are represented as means + SD (error bars) of three independent experiments.
Kinetics of viral infection
To determine if the swine tracheal explants supported productive viral replication, explants were infected with 2·5 × 102 pfu of swine influenza virus and maintained in organ culture for 5 days. Infectious extracellular virus extracted from the epithelial surface was quantified in plaque assays in MDCK cells. A marked increase in the amount of infectious virus was observed 24 hours pi, when the number of infectious units increased by approximately 10‐fold when compared with virus recovered immediately after infection. Viral titres consistently increased by 3 log10 on day 2 and remained stable up to day 4 pi (at c 6·4 log10) followed by a small decline by day 5 pi (Figure 5E), showing that tracheal explants support productive viral infection.
A qPCR assay to measure intracellular and extracellular virus was used as a complementary method to assess infection kinetics. To this end, explants were inoculated with 2·5 × 101, 2·5 × 102 or 2·5 × 104 pfu, and maintained in organ culture for 5 days. Viral quantification showed that infection kinetics was dose dependent (Figure 5F–H). In explants inoculated with 2·5 × 104 pfu, the peak viral load was observed at day 1 pi. This was reduced at day 3 pi but then there followed a resurgence in copy number by day 5 pi (Figure 5F). Inoculation with 2·5 × 102 pfu showed a different pattern of infection in which the peak shifted to day 3 pi, with a higher viral copy number than the one observed when a higher dose was used (Figure 5G). Viral RNA was detected at day 2 pi in explants inoculated with the lowest dose (2·5 × 101 pfu) and a marked increase in viral copy number was observed at days 4 and 5 pi (Figure 5H).
Discussion
We have established an ex vivo air interface tracheal organ culture for the study of influenza viruses which could also be used to study other respiratory pathogens. EVOC systems have been developed previously for the study of respiratory pathogens using human, swine, bovine, canine and horse respiratory tissue. 18 , 20 , 23 , 25 , 27 , 29 , 30 The experimental value of the tracheal explants described here is illustrated by the preservation of good levels of both histological and functional features for seven consecutive days, and by showing productive infection with an avian‐like swine H1N1 virus.
A vast body of knowledge in influenza biology has been generated using diverse experimental systems, each possessing advantages and disadvantages. In vivo studies have provided valuable data on influenza pathogenesis, although detailed information about quantitative infection dynamics and the host innate immune response is still scarce. 1 , 3 , 4 , 5 , 15 , 37 , 38 For respiratory pathogens in particular, samples are usually collected by nasal swabs or bronchoalveolar lavage, which may not fully reflect the tissue‐specific bacterial and viral infection dynamics occurring throughout the respiratory tract, making it difficult to dissect regional tissue‐specific effects from generalized systemic responses. Cell culture systems, using epithelial cells from the respiratory mucosa, may lack key physiological features such as cell polarization, reliable expression levels of cellular surface receptors or the intricate relationships between the different cell types present in the respiratory tract. 39 Primary cell cultures are difficult to establish, possess an inadequate distribution of the different cell types and become altered after a limited number of passages. Established continuous cell lines are by definition aberrant and usually belong to a cell type different to the natural target cells and, in some cases, are derived from a species different from the natural host. Ex vivo organ cultures provide an alternative solution. Nasal mucosa 27 and tracheal explants 39 immersed in cell culture medium have been used previously for the study of influenza infection. However, using this technique cells are subjected to submersion‐related stress and to a continuous source of infecting pathogen as the inoculum remains in the cell culture medium. 22
The tracheal organ culture described here exhibits several advantages compared with submerged organ cultures, cell cultures or in vivo studies. First, in an air interface system the explants are exposed basolaterally to culture medium and the apical cell surface is exposed to the air. The three‐dimensional structure of the tissue as well as the interactions among cells is preserved. In particular, the cells of the epithelial lining of the trachea maintain the same morphological appearance, differentiation and polarity as described in the living host. In contrast to submerged systems, once explants are inoculated, re‐infection from the culture medium or other components of the system is rare. The absence of a blood supply constitutes a weakness of the air interface EVOC system as recruitment of cells of the immune system to the infection site is abrogated. However, such a potential pitfall might be turned to advantage and used to dissect the effects of innate and intrinsic immunity. 40
In this study, the overall histology of the explants was preserved for 7 days with no significant differences in the thickness of the uninfected epithelium. This is consistent with previous reports of similar systems in which explants of respiratory tissue from different species have been maintained for variable periods of time, ranging from 60 hours to 5 days. 23 , 25 , 27
Mucus production provides an important nonspecific defence against viral infections. 41 , 42 During the initial washing steps in setting up the explants, a majority of the mucus is removed from the epithelial surface. However, mucus is produced de novo by goblet cells throughout the duration of the organ culture, and thus the physiological conditions present in the living host are maintained in this regard.
We also evaluated the function of the mucociliary escalator system, and thus also of co‐ordinated ciliary function of the tracheal explants, using a simple bead clearance assay. For beads to be cleared from the surface of the organ culture piece, individual cilia must beat in a coordinated manner across the epithelial surface with mucus acting as a vehicle to transport unwanted particles away from where the alveoli would have been in a wave‐like motion towards where the larynx would have been. 43 By using the bead clearance assay, we were able to distinguish between small patches of ciliary movement and truly coordinated ciliary activity, an important measure of the robustness of the system. The increase in the clearance time observed at later time points in uninfected explants may be associated with changes in pH, or energy limitations of the tissue. Although the supplemented culture medium contains most of the essential components required for survival of the explants, it may not constitute a complete substitute for the nutrients available from the living host.
Infection of swine tracheal explants with A/swine/England/435/06 (H1N1) resulted in productive infection with histopathological and immunohistochemical patterns consistent with those observed in influenza infections of both humans and pigs. 3 , 4 , 13 , 36 As expected, infection kinetics was dose dependent: when using a high dose of virus we observed a mild infection that peaked bimodally at day 1 and day 5 pi. With an intermediate dose, a single peak occurred at day 3 pi, whereas the lowest infection dose resulted in detectable virus at day 2 pi, followed by a subsequent rise in copy number at later time points. Such distinct patterns might be related to differential levels of innate immune response, as well as to the initial number of susceptible target cells. A large inoculum would be expected to infect most of the susceptible cells and viral replication may also trigger a massive innate immune response, resulting in a small number of susceptible cells available for a second round of viral replication. Once the antiviral signalling molecules are depleted, the remaining infectious particles can infect the susceptible cells, and this would be consistent with the second rise in virus titres observed at day 5 (Figure 5F). Although a second round of infection could not be ruled out, the peaks are too separated in time for this to be likely. A smaller dose of virus might trigger a milder innate immune response, restricted to neighbouring cells, allowing the virus to infect a larger number of cells over multiple rounds of infection. Accordingly, the kinetics of infection with the lowest viral dose displayed a marked shift to the right because more replication cycles were required to infect all or most of the susceptible cells. Further experiments that incorporate the quantification of cytokine expression will help to test this hypothesis.
In our study, an influenza virus of swine origin was used in the validation of the pig trachea EVOC system. Future studies, including viruses of different subtypes isolated from diverse hosts, should be performed to evaluate the susceptibility of pig trachea to influenza infection and to study the mechanisms underlying any possible host‐specific specificity.
In summary, the EVOC system constitutes a useful biological tool with a wide range of possible applications for the study of influenza infection, including the evaluation of mutants obtained by reverse genetics, the study of natural variability and reassortment events, the effects of antiviral drugs in natural target cells, investigation of host susceptibility and range of influenza viruses and the development of mathematical models of infection dynamics. When possible, EVOC systems should be considered as an alternative to in vivo experiments, applying the principles of 3Rs in animal experimentation.
Acknowledgements
We thank Yvonne Spencer, Sara Marsh, Alejandro Nunez (VLA‐Weybridge) and Carina Ferrari for their assistance with the immunohistochemistry protocol and also T. J. McKinley for his assistance in the statistical analysis of data. This work was supported by a grant from Defra and Hefce under the Veterinary Training and Research Initiative to the Cambridge Infectious Diseases Consortium (CIDC) through a fellowship awarded to S. F. Nunes. J.L.N.W. is funded by the Alborada Trust and P.R.M. is funded by the Wellcome Trust. The swine influenza virus was obtained through a programme of influenza surveillance in GB pigs funded by Defra (project ED1204).
References
- 1. Webster RG, Bean WJ, Gorman OT, Chambers TM, Kawaoka Y. Evolution and ecology of influenza A viruses. Microbiol Rev 1992; 56:152–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Fouchier RAM, Munster V, Wallensten A et al. Characterization of a novel influenza A virus hemagglutinin subtype (H16) obtained from black‐headed gulls. J Virol 2005; 79:2814–2822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Brown IH. The epidemiology and evolution of influenza viruses in pigs. Vet Microbiol 2000; 74:29–46. [DOI] [PubMed] [Google Scholar]
- 4. Taubenberger JK, Morens DM. The pathology of influenza virus infections. Annu Rev Pathol 2008; 3:499–522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Maines TR, Szretter KJ, Perrone L et al. Pathogenesis of emerging avian influenza viruses in mammals and the host innate immune response. Immunol Rev 2008; 225:68–84. [DOI] [PubMed] [Google Scholar]
- 6. Ito T, Couceiro JN, Kelm S et al. Molecular basis for the generation in pigs of influenza A viruses with pandemic potential. J Virol 1998; 72:7367–7373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kida H, Ito T, Yasuda J et al. Potential for transmission of avian influenza viruses to pigs. J Gen Virol 1994; 75 (Pt 9):2183–2188. [DOI] [PubMed] [Google Scholar]
- 8. Scholtissek C, Bürger H, Kistner O, Shortridge KF. The nucleoprotein as a possible major factor in determining host specificity of influenza H3N2 viruses. Virology 1985; 147:287–294. [DOI] [PubMed] [Google Scholar]
- 9. Shinde V, Bridges CB, Uyeki TM et al. Triple‐reassortant swine influenza A (H1) in humans in the United States, 2005–2009. N Engl J Med 2009; 360: 2616–25. [DOI] [PubMed] [Google Scholar]
- 10. Garten RJ, Davis CT, Russell CA et al. Antigenic and genetic characteristics of swine‐origin 2009 A(H1N1) influenza viruses circulating in humans. Science 2009; 325: 197–201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barnard DL. Animal models for the study of influenza pathogenesis and therapy. Antiviral Res 2009; 82:A110–A122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Van Reeth K, Van Gucht S, Pensaert M. Correlations between lung proinflammatory cytokine levels, virus replication, and disease after swine influenza virus challenge of vaccination‐immune pigs. Viral Immunol 2002; 15:583–594. [DOI] [PubMed] [Google Scholar]
- 13. Van Reeth K. Cytokines in the pathogenesis of influenza. Vet Microbiol 2000; 74:109–116. [DOI] [PubMed] [Google Scholar]
- 14. Van Reeth K, Van Gucht S, Pensaert M. In vivo studies on cytokine involvement during acute viral respiratory disease of swine: troublesome but rewarding. Vet Immunol Immunopathol 2002; 87:161–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Zambon MC. The pathogenesis of influenza in humans. Rev Med Virol 2001; 11:227–241. [DOI] [PubMed] [Google Scholar]
- 16. Thompson CI, Barclay WS, Zambon MC, Pickles RJ. Infection of human airway epithelium by human and avian strains of influenza A virus. J Virol 2006; 80:8060–8068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Daidoji T, Koma T, Du A et al. H5N1 avian influenza virus induces apoptotic cell death in mammalian airway epithelial cells. J Virol 2008; 82:11294–11307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anderton TL, Maskell DJ, Preston A. Ciliostasis is a key early event during colonization of canine tracheal tissue by Bordetella bronchiseptica . Microbiology 2004; 150:2843–2855. [DOI] [PubMed] [Google Scholar]
- 19. Dugal F, Girard C, Jacques M. Adherence of Bordetella bronchiseptica 276 to porcine trachea maintained in organ culture. Appl Environ Microbiol 1990; 56:1523–1529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Jackson AD, Rayner CF, Dewar A, Cole PJ, Wilson R. A human respiratory‐tissue organ culture incorporating an air interface. Am J Respir Crit Care Med 1996; 153:1130–1135. [DOI] [PubMed] [Google Scholar]
- 21. Jang YJ, Lee SH, Kwon H‐J, Chung Y‐S, Lee B‐J. Development of rhinovirus study model using organ culture of turbinate mucosa. J Virol Methods 2005; 125:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Leeming G, Meli ML, Cripps P et al. Tracheal organ cultures as a useful tool to study Felid herpesvirus 1 infection in respiratory epithelium. J Virol Methods 2006; 138:191–195. [DOI] [PubMed] [Google Scholar]
- 23. Priestnall SL, Mitchell JA, Brooks HW, Brownlie J, Erles K. Quantification of mRNA encoding cytokines and chemokines and assessment of ciliary function in canine tracheal epithelium during infection with canine respiratory coronavirus (CRCoV). Vet Immunol Immunopathol 2009; 127:38–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Rayner CF, Jackson AD, Rutman A et al. Interaction of pneumolysin‐sufficient and ‐deficient isogenic variants of Streptococcus pneumoniae with human respiratory mucosa. Infect Immun 1995; 63:442–447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Soane MC, Jackson A, Maskell D et al. Interaction of Bordetella periitussis with human respiratory mucosa in vitro. Respir Med 2000; 94:791–799. [DOI] [PubMed] [Google Scholar]
- 26. Tsang KW, Rutman A, Tanaka E et al. Interaction of Pseudomonas aeruginosa with human respiratory mucosa in vitro. Eur Respir J 1994; 7:1746–1753. [DOI] [PubMed] [Google Scholar]
- 27. Glorieux S, Van den Broeck W, Van Der Meulen KM, Van Reeth K, Favoreel HW, Nauwynck HJ. In vitro culture of porcine respiratory nasal mucosa explants for studying the interaction of porcine viruses with the respiratory tract. J Virol Methods 2007; 142:105–112. [DOI] [PubMed] [Google Scholar]
- 28. Vandekerckhove A, Glorieux S, Broeck WV, Gryspeerdt A, Van Der Meulen KM, Nauwynck HJ. In vitro culture of equine respiratory mucosa explants. Vet J 2009; 181:280–287. [DOI] [PubMed] [Google Scholar]
- 29. Niesalla HS, Dale A, Slater JD et al. Critical assessment of an in vitro bovine respiratory organ culture system: a model of bovine herpesvirus‐1 infection. J Virol Methods 2009; 158:123–129. [DOI] [PubMed] [Google Scholar]
- 30. Hamilton A, Robinson C, Sutcliffe IC et al. Mutation of the maturase lipoprotein attenuates the virulence of Streptococcus equi to a greater extent than does loss of general lipoprotein lipidation. Infect Immun 2006; 74:6907–6919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Dimova S, Brewster ME, Noppe M, Jorissen M, Augustijns P. The use of human nasal in vitro cell systems during drug discovery and development. Toxicol In Vitro 2005; 19:107–122. [DOI] [PubMed] [Google Scholar]
- 32. Russell WMS, Burch RL. The Principles of Humane Experimental Technique. London: Methuen, 1959. [Google Scholar]
- 33. Matrosovich M, Matrosovich T, Garten W, Klenk HD. New low‐viscosity overlay medium for viral plaque assays. Virol J 2006; 3:63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hoffmann E, Stech J, Guan Y, Webster RG, Perez DR. Universal primer set for the full‐length amplification of all influenza A viruses. Arch Virol 2001; 146:2275–2289. [DOI] [PubMed] [Google Scholar]
- 35. Spackman E, Senne DA, Myers TJ et al. Development of a real‐time reverse transcriptase PCR assay for type A influenza virus and the avian H5 and H7 hemagglutinin subtypes. J Clin Microbiol 2002; 40:3256–3260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Jung T, Choi C, Chae C. Localization of swine influenza virus in naturally infected pigs. Vet Pathol 2002; 39:10–16. [DOI] [PubMed] [Google Scholar]
- 37. Baigent SJ, McCauley JW. Influenza type A in humans, mammals and birds: determinants of virus virulence, host‐range and interspecies transmission. Bioessays 2003; 25:657–671. [DOI] [PubMed] [Google Scholar]
- 38. Hilleman MR. Realities and enigmas of human viral influenza: pathogenesis, epidemiology and control. Vaccine 2002; 20:3068–3087. [DOI] [PubMed] [Google Scholar]
- 39. Lin C, Holland RE Jr, Williams NM, Chambers TM. Cultures of equine respiratory epithelial cells and organ explants as tools for the study of equine influenza virus infection. Arch Virol 2001; 146:2239–2247. [DOI] [PubMed] [Google Scholar]
- 40. Bieniasz PD. Intrinsic immunity: a front‐line defense against viral attack. Nat Immunol 2004; 5:1109–1115. [DOI] [PubMed] [Google Scholar]
- 41. Tamura S, Kurata T. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn J Infect Dis 2004; 57:236–247. [PubMed] [Google Scholar]
- 42. Randell SH, Boucher RC. Effective mucus clearance is essential for respiratory health. Am J Respir Cell Mol Biol 2006; 35:20–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Braiman A, Priel Z. Efficient mucociliary transport relies on efficient regulation of ciliary beating. Respir Physiol Neurobiol 2008; 163:202–207. [DOI] [PubMed] [Google Scholar]


