Abstract
Fecal transplants are increasingly utilized for treatment of recurrent infections (i.e., Clostridium difficile) in the human gut and as a general research tool for gain-of-function experiments (i.e., gavage of fecal pellets) in animal models. Changes observed in the recipient's biology are routinely attributed to bacterial cells in the donor feces (~1011 per gram of human wet stool). Here, we examine the literature and summarize findings on the composition of fecal matter in order to raise cautiously the profile of its multipart nature. In addition to viable bacteria, which may make up a small fraction of total fecal matter, other components in unprocessed human feces include colonocytes (~107 per gram of wet stool), archaea (~108 per gram of wet stool), viruses (~108 per gram of wet stool), fungi (~106 per gram of wet stool), protists, and metabolites. Thus, while speculative at this point and contingent on the transplant procedure and study system, nonbacterial matter could contribute to changes in the recipient's biology. There is a cautious need for continued reductionism to separate out the effects and interactions of each component.
Fecal transfers are increasingly common in animal models and humans. This essay notes that bacteria may not be the only player in donor feces that can affect the recipient's biology.
Introduction
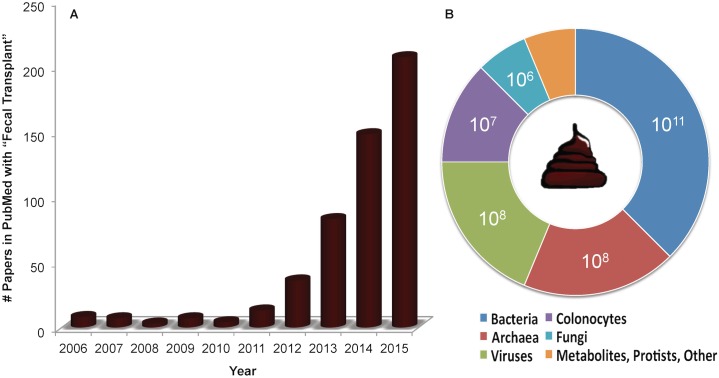
A fecal transplant—the transfer of stool or portions of stool from one organism into the gastrointestinal tract of another—is rapidly gaining attention as a treatment for human gut infections and as a tool for functional "knock-in" studies of the microbiota in animal models. In humans, the procedure is referred to as fecal microbiota transplantation because the microbial components are typically enriched, and in animal models, the transfer of unprocessed stool is commonly achieved by feeding or oral gavage of fecal matter. For the purposes of this essay, we will use the catch-all phrase of “fecal transplants” to refer to all types of procedures. Fig 1 shows the very recent growth of the term in PubMed references involving both human and model system studies.
Fig 1. The growth of fecal transplants as reflected in references in PubMed and the estimated composition of human feces.
The charts show (A) the rapid rise in publications on fecal transplants in the National Library of Medicine's search service (PubMed), particularly between 2012 and 2015, and (B) the estimated upper concentration of the biological entity per gram of unprocessed human feces, as cited in the text. Estimates do not necessarily reflect the viable number of the biological entity, and the concentration of the archaea is estimated from a methanogen breath test that is not solely based on the presence of archaea. Concentrations of metabolites, protists, and other entities were not identified.
Several analyses report clinical resolution of Clostridium difficile infection (CDI) [1–5], though the long-term effects of the transplants are unknown [6]. Preliminary results also demonstrate positive outcomes for insulin sensitivity [7], multiple sclerosis [8], and Crohn’s disease [9]. The presumptive element connecting these conditions is the gut’s bacterial community, and thus the treatment’s success enthusiastically revolves around intestinal bacteria that are assumed mostly viable in feces. There are a few studies in mice and humans that validate the positive effects of cultured bacteria on CDI [10,11] and mucosal barrier function [12]. Additionally, the microbial portion of human stool can be highly enriched from other fecal material through microfiltration [13,14], spore fractionation [15], and density gradients [16].
Here, we tentatively emphasize that viable bacteria may not be the only player in donor feces that affect the recipient's biology, a fact that is well appreciated by experts. Viruses, archaea, fungi, animal colonocytes, protists, and a number of metabolites that commensal bacteria make or are dependent upon can potentially occur in unprocessed feces. Here, we accentuate the patterns seen in fecal composition analyses and various experiments that illuminate functional effects of individual components of fecal matter. We also highlight important and tractable questions for which further reductionism could help deconstruct the benefit of constituent parts of fecal matter.
Fecal Composition
Human fecal composition has not been intensively studied. The studies that have examined composition are mostly from the 1970s and 1980s and report varying results, perhaps because of variation in diet and health. On average, adult fecal matter is estimated to be 75% water and 25% solid matter [17]. The vast majority of solid matter is organic material, whose makeup consists of 25%–54% microbial cells (with a slight portion likely consisting of viruses) that may be alive or dead [18]. As microbial counts were based on light microscopy and a modification of the Gram stain, the microbial cells were presumed to be mostly bacteria [18], but quality evidence is lacking. Several other components are found in significant concentration, including archaea, fungi, and microbial eukaryotes. One particular methanoarchaeon species, Methanobrevibacter smithii, was detected in 95.7% of patients spanning infants, adults, and the elderly [19], and it can comprise up to 10% of all fecal anaerobes [20]. Viable colonocytes are also readily isolated from newborn and adult feces [21–23]. No analysis of their potential contribution to the success of fecal transplants has been reported. Independent validations of these estimates are needed, particularly measurements that consider all of the entities at once.
While transplants can be highly effective treatment in certain cases, concerns remain about the hypothetical co-transfer of pathogenic microbes [24]. Contamination by environmental microbes is also a risk during the collection, storage, and handling of donor stool, as seen in the early periods of blood storage for transfusions [25,26]. To standardize laboratory protocols and enhance stability of fecal matter, one option is to use frozen donor material from rigorously screened volunteers. Several studies compared the efficacy of frozen versus fresh stool on recurrent or refractory CDI and reported little to no difference [14,27,28]. Extensive longitudinal screening of stool donors is essential to track the long-term success of treatment, and further metagenomic studies of the transferred fecal material and transfer proficiency to recipients are warranted.
Bacteria
It is well established that the gut contains the highest density of microbes in the human body, with the bacteria-to-human cell ratio recently estimated to be 1.3:1 [29]. In feces, bacteria constitute 25%–54% of solid matter [18] and thus between 6.3% and 13.5% of total fecal matter. Averaged estimates from 14 studies yield a mean bacterial concentration of nearly 1011 bacteria per gram of wet stool [29]. Yet, a clear distinction was shown in a study between the viable (49%), injured (19%), and dead (32%) bacterial cells collected from fresh fecal samples under anaerobic conditions [30]. These statistics indicate that only 3.0%–6.6% of total fecal matter may be composed of viable bacteria. The percentage could conceivably be even lower if samples are handled in aerobic conditions for lengthy amounts of time, although frequent aerobic preparation of fecal material has resulted in high cure rates. Furthermore, and as previously noted, transplants with frozen fecal samples that may have reduced viable bacteria can lead to an almost identical resolution of CDI to transplants with fresh samples [27,31]. It should be noted, however, that even bacterial DNA or dead cells might retain some immunostimulatory functions, as colitis symptoms in a dextrose sodium sulfate–induced mouse model were strikingly alleviated by introduction of probiotic DNA and unviable irradiated bacterial cells [32,33].
Other studies suggest that interactions between the host genotype and microbiota can potentially affect transplant outcomes. Across a collection of studies, human fecal donations from related donors showed slightly higher resolution in CDI cases (93%) compared to unrelated donors (84%) [34]. This observation is notable in light of the recent finding that human genetic variation is significantly correlated with variation in bacterial community composition [35,36]. However, a recent meta-analysis demonstrated no significant difference in efficacy between related and unrelated donors [37]. Furthermore, a placebo-controlled trial resulted in the successful treatment of seven of nine people who received a transplant from a single, unrelated, donor [38]. Thus, the evidence to date suggests that relatedness either has little or no effect on treating CDI.
To demonstrate that bacteria directly contribute to disease resolution, several research groups have tested whether enriched bacterial portions of fecal material can be effective in treating CDI in mice and humans. Use of a six-species cocktail therapy suppressed recurrent CDI in 92% of mice [10] when approximately 1010 cells per bacterial species were gavaged into recipients. In another mouse study, 108 colony-forming units of a single bacterium isolate, Lachnospiraceae D4, caused over a 10-fold reduction in the number of C. difficile colony-forming units per gram of cecal contents [39]. A cocktail of nontoxigenic C. difficile spores was also successfully used in suppressing CDI recurrence in a human trial [40]. At 26 weeks of treatment, only 0%–5% of patients from various treatment groups had toxigenic C. difficile remaining in feces. These studies indicate that cultured bacteria can, in certain cases, be effective contributors to CDI disease resolution.
Viruses
Viruses from eukaryotes, bacteria, and archaea are less studied components of the gut microbiota than bacteria. From five fecal samples, count estimates indicate that the viral abundance ranges from 108 to 109 viruses per gram of feces (wet weight), and the average virus-to-bacterium ratio is 0.13 [41]. These estimates are comparably low to those reported in other environments where the virus-to-microbial cell ratios range from 1.4 to 160 [42], which supports the emerging view that viruses exhibit a more temperate lifestyle in the gut [43,44]. Additionally, a recent metagenomic study demonstrated that numerous temperate phages are transferred during fecal transplants [24]. Prophages often assist in controlling invading pathogens, modulating community structure, and maintaining gut homeostasis [44]. The dominance of temperate viruses is, however, typical of healthy control feces, as patients suffering from bowel diseases can have increased amounts of virulent phages [45]. One of the most abundant, conserved, and prevalent bacteriophages in the human gut is crAssphage [46], a finding that suggests some phages may be highly conserved in the human population.
The impact of bacteriophages on human health is under active consideration. Phage therapy entails the isolation and inoculation of phages (or their antibacterial enzymes) that target a specific bacterium. While not all phage treatments are effective [47], several in vitro and in vivo experiments have been successful. As a treatment for CDI, 108 plaque-forming units per mL of a specific phage were introduced into a human colon model. Over a period of 35 days, the treatment caused a significant decrease in vegetative C. difficile cells (albeit there was an increase in C. difficile spores) as well as toxin production to levels below the detection threshold of the assay [48]. Control replicates contained high concentrations of both vegetative cells and toxin. Phage therapy of CDI in a hamster model also significantly delayed bacterial colonization and the onset of symptoms [49]. Specific phage cocktails could, in theory, allow commensal bacteria that are in competition with C. difficile to reflourish in the gut [50]. While C. difficile phages may eventually be developed into therapeutic agents, there is yet no evidence that phages specific to C. difficile are transferred in fecal transplants.
There have been several concerns about the safety of phage therapy. To alleviate the apprehension, a recent human clinical trial orally inoculated a group of 15 subjects with a high dose of 17 phage groups targeting Escherichia coli and Proteus infection and found no adverse effects [51]. Phage therapy cocktails have continuously demonstrated potential to target and eliminate specific virulent bacteria while avoiding adverse effects typical of antibiotics (e.g., resistance, diarrhea, etc.) [52–54]. However, a potential drawback is the risk of evolution of bacterial resistance to phages [55,56], though phages can potentially evolve counter-resistance mechanisms. Furthermore, human studies involving phage therapy are relatively small-scale thus far. Larger patient cohorts and further studies of phage dosages, evolution of phage host ranges and bacterial resistance [56], and the stability of phage-based drugs are needed.
Archaea and Fungi
Archaea are well-recognized but relatively understudied members of the human gut microbiota [57], with methanoarchaeon comprising up to 10% of fecal anaerobes [20]. Based on the concentration of methane in breath, estimates suggest a minimum presence of 107–108 methanogens per gram of both dry and wet stool [58,59], though it is unresolved what percentage of these methanogens are from archaea versus bacteria. Higher than normal concentrations of intestinal archaea are associated with Crohn’s disease and multiple sclerosis [60]. Similarly, fungi in the gut have been cultured in 70% of healthy adults [61]. They occur in estimated concentrations of up to 106 microorganisms per gram of feces [62] and appear to comprise only 0.03% of all microbes in feces [63]. Candida albicans is the most common and studied yeast, but it is kept in check by competitive commensal bacteria in a healthy gut. When bacterial homeostasis is disturbed, however, C. albicans increases its numbers drastically [64,65]. These fungi may also help induce intestinal diseases by penetrating the intestinal colonocyte barrier and driving inflammation [66]. Indeed, high concentrations of C. albicans occur in individuals with inflammatory bowel diseases [67,68]. The contribution of archaea and fungi to changes in function will be an important area of future research.
Human Colonocytes
Interestingly, viable epithelial cells of the large intestine, or colonocytes, can be isolated at a concentration of up to 107 per gram of wet fecal material [23]. Viable colonic cells have effectively been isolated from newborn fecal samples (>80% viable) [21] and biopsy specimens from colonic crypts (>98% viable) [22]. Isolation is possible due to the resilient ability of colonocytes to take on a globular shape and survive once exfoliated into the fecal stream [69]. Thus, their viability and partial functionality is likely retained in the course of some transplant treatments, especially in animal models that utilize feeding or oral gavage of fecal material.
By acting as the physical barrier between bacteria and the host’s internal tissues and organs, colonocytes allow host tolerance of the intestinal microbiota [70]. When high levels of colonocyte death occur, their mediating role disintegrates because of increased intestinal permeability [71]. Indeed, major pathological conditions of the bowel are associated with changes in the growth and functions of the colonic epithelium [22,72], similar to changes frequently observed in microbiota studies. Their restoration is key in successful recovery from such conditions. A recent study transplanted healthy viable colon stem cells into an immunodeficient mouse model with superficial colon damage and found that cells readily integrated, and a single layer of epithelium fully covered areas lacking colonocytes [73]. The presence of colonic stem cells in feces has yet to be recorded, although one study recovered stem cells from the colonic epithelium that often sheds into the fecal stream [74]. Should colonic stem cells be identified in feces in human or animal models, they may affect the success of transplants if they can engraft in recipients.
In addition to colonocytes, molecules such as immunoglobulin A (IgA) can act as the first line of defense for the intestinal epithelium [75]. IgA reinforces the intestinal barrier and protects host cells against pathogens and enteric toxins in the gut [75]. For instance, IgA significantly inhibited C. difficile toxin binding to hamster intestinal brush border membranes compared to the control [76]. Likewise, human epithelial cell lines with IgA added to their surface showed a decrease in C. difficile–associated pathology compared to cells lacking IgA [77]. It remains to be seen if introducing IgA directly into human subjects will be beneficial.
Metabolites
It is well known that fiber is metabolized by intestinal bacteria to produce short-chain fatty acids (SCFA) that have prominent anti-inflammatory and T cell–inducing properties in the colon [78–80]. Fiber strongly contributes to fecal weight, and low fiber diets in mice can lead to an irreversible loss in bacterial diversity [81]. While direct reintroduction of missing fiber in this study did not restore the diversity, transplants from mice with a high fiber diet did. Furthermore, low fiber diets lead to “microbial starving,” whereby once-commensal bacteria attack the intestinal lining [82]. Fiber supplements used in a study with C. difficile–infected hamsters, however, managed to significantly modulate onset time of systemic symptoms [83]. Fiber intake has also been linked to increased microbial diversity and reduced obesity in humans [84,85].
Butyrate-producing bacteria or butyrate concentrations in feces can be lower in patients with colorectal cancer and ulcerative colitis [86–88]. Preliminary studies of enemas with butyrate or SCFA cocktails (acetate, butyrate, and propionate) show some resolution in patients with distal ulcerative colitis [89–93]. Following these treatments, 35%–67% of patients exhibited improvement. Furthermore, oral administration of sodium butyrate in a colitis mouse model alleviated inflammation and mucosal damage [94], and propionate led to improvement of symptoms in a multiple sclerosis mouse model by promoting regulatory T cell differentiation [95]. No adverse side effects were noted in any of these studies, though some metabolite enemas are malodorous. One review, however, cautions against the use of such metabolites [96]. While butyrate acts as an energy source, increases colonocyte growth, and decreases apoptosis of colonocytes under healthy conditions [97], excess butyrate accumulation around human colonic carcinoma cells has been connected with increased apoptosis [98]. Finally, estimates suggest there are nearly 900 gene clusters in human gut–associated bacteria that make small molecules [99]. Determining functions may be important in understanding the composite nature of feces and its effects on fecal transplants in humans and/or animal models.
Summary
Here, we cautiously note that bacteria, either viable or unviable in transferred fecal material, may not be the only player in donor feces that affects the recipient's biology. On the one hand, the effects of bacteria on CDI or animal model traits such as obesity [100] and toxin tolerance [101] appear well justified thus far. On the other hand, in a broader context where fecal transplants are solely utilized in animal model studies and other human diseases, judicious reductionism seems warranted in light of a limited understanding of the complex nature of feces. Deconstructing the benefit and interactions of constituent parts of fecal matter will clarify the relative importance and causality of each of these components and the potential development of specific therapies.
Key Points and Future Directions
A few studies using cocktails of bacteria in animal models and humans show suppression of CDI. However, these studies are preliminary and limited.
Through bacterial targeting, phage therapy can potentially eliminate virulent bacteria in a diseased gut and allow commensal bacterial to reflourish.
Colonocytes prevent bacterial translocation into internal tissues and organs; transplants of healthy viable colon stem cells into mouse models result in repair of superficial colon damage.
Metabolites can nourish the colonocyte barrier and intestinal bacteria. Oral administration of metabolites can alleviate inflammation, mucosal damage, and multiple sclerosis symptoms. However, only 5% of the SCFAs produced in the distal colon are estimated to be excreted into feces [102]. Hence, metabolite concentrations are likely to be much lower than concentrations used in oral administration studies.
Archaea and fungi are common in feces. Though high concentrations of intestinal archaea and certain fungi have been correlated to both intestinal and autoimmune diseases, their causative effects are unknown.
Human genetic relatedness has little to no influence on the effectiveness of human fecal transplants, though genetic factors do shape bacterial community composition.
Individual components of fecal matter can yield health benefits and may work synergistically to restore homeostasis. There is a cautious need for continued reductionism to understand the precise benefit and interactions of various fecal components.
Acknowledgments
We thank Sean Davies, Ken Lau, Vincent Young, Kevin Kohl, Mike Sadowski, and Joseph Zackular for providing helpful feedback on the manuscript.
Abbreviations
- CDI
Clostridium difficile infection
- IgA
immunoglobulin A
- SCFA
short-chain fatty acids
Funding Statement
This work was supported by National Science Foundation Awards DEB 1046149 and IOS 1456778. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Kassam Z, Hundal R, Marshall JK, Lee CH (2012) Fecal transplant via retention enema for refractory or recurrent Clostridium difficile infection. Archives of Internal Medicine 172: 191–193. 10.1001/archinte.172.2.191 [DOI] [PubMed] [Google Scholar]
- 2.van Nood E, Vrieze A, Nieuwdorp M, Fuentes S, Zoetendal EG, et al. (2013) Duodenal infusion of donor feces for recurrent Clostridium difficile. N Engl J Med 368: 407–415. 10.1056/NEJMoa1205037 [DOI] [PubMed] [Google Scholar]
- 3.Yoon SS, Brandt LJ (2010) Treatment of refractory/recurrent C. difficile-associated disease by donated stool transplanted via colonoscopy: a case series of 12 patients. J Clin Gastroenterol 44: 562–566. 10.1097/MCG.0b013e3181dac035 [DOI] [PubMed] [Google Scholar]
- 4.Aas J, Gessert CE, Bakken JS (2003) Recurrent Clostridium difficile colitis: case series involving 18 patients treated with donor stool administered via a nasogastric tube. Clin Infect Dis 36: 580–585. [DOI] [PubMed] [Google Scholar]
- 5.Rao K, Young VB (2015) Fecal microbiota transplantation for the management of Clostridium difficile infection. Infect Dis Clin North Am 29: 109–122. 10.1016/j.idc.2014.11.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borody TJ, Khoruts A (2012) Fecal microbiota transplantation and emerging applications. Nat Rev Gastroenterol Hepatol 9: 88–96. [DOI] [PubMed] [Google Scholar]
- 7.Vrieze A, Van Nood E, Holleman F, Salojarvi J, Kootte RS, et al. (2012) Transfer of intestinal microbiota from lean donors increases insulin sensitivity in individuals with metabolic syndrome. Gastroenterology 143: 913–916. e917 10.1053/j.gastro.2012.06.031 [DOI] [PubMed] [Google Scholar]
- 8.Borody TJ, Leis S, Campbell J, al. e (2011) Fecal microbiota transplantation (FMT) in multiple sclerosis (MS) [abstract]. Am J Gastroenterol 106:S352. [Google Scholar]
- 9.Suskind DL, Brittnacher MJ, Wahbeh G, Shaffer ML, Hayden HS, et al. (2015) Fecal microbial transplant effect on clinical outcomes and fecal microbiome in active Crohn's disease. Inflamm Bowel Dis 21: 556–563. 10.1097/MIB.0000000000000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lawley TD, Clare S, Walker AW, Stares MD, Connor TR, et al. (2012) Targeted restoration of the intestinal microbiota with a simple, defined bacteriotherapy resolves relapsing Clostridium difficile disease in mice. PLoS Pathog 8: e1002995 10.1371/journal.ppat.1002995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Petrof EO, Gloor GB, Vanner SJ, Weese SJ, Carter D, et al. (2013) Stool substitute transplant therapy for the eradication of Clostridium difficile infection: 'RePOOPulating' the gut. Microbiome 1:3 10.1186/2049-2618-1-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li M, Liang P, Li Z, Wang Y, Zhang G, et al. (2015) Fecal microbiota transplantation and bacterial consortium transplantation have comparable effects on the re-establishment of mucosal barrier function in mice with intestinal dysbiosis. Front Microbiol 6:692 10.3389/fmicb.2015.00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hamilton MJ, Weingarden AR, Unno T, Khoruts A, Sadowsky MJ (2013) High-throughput DNA sequence analysis reveals stable engraftment of gut microbiota following transplantation of previously frozen fecal bacteria. Gut Microbes 4: 125–135. 10.4161/gmic.23571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hamilton MJ, Weingarden AR, Sadowsky MJ, Khoruts A (2012) Standardized frozen preparation for transplantation of fecal microbiota for recurrent Clostridium difficile infection. Am J Gastroenterol 107: 761–767. 10.1038/ajg.2011.482 [DOI] [PubMed] [Google Scholar]
- 15.Khanna S, Pardi DS, Kelly CR, Kraft CS, Dhere T, et al. (2016) A Novel Microbiome Therapeutic Increases Gut Microbial Diversity and Prevents Recurrent Clostridium difficile Infection. The Journal of Infectious Diseases 10.1093/infdis/jiv766 [DOI] [PubMed] [Google Scholar]
- 16.Hevia A, Delgado S, Margolles A, Sánchez B (2015) Application of density gradient for the isolation of the fecal microbial stool component and the potential use thereof. Scientific Reports 5: 16807 10.1038/srep16807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rose C, Parker A, Jefferson B, Cartmell E (2015) The Characterization of Feces and Urine: A Review of the Literature to Inform Advanced Treatment Technology. Crit Rev Environ Sci Technol 45: 1827–1879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stephen AM, Cummings JH (1980) The microbial contribution to human faecal mass. J Med Microbiol 13: 45–56. [DOI] [PubMed] [Google Scholar]
- 19.Dridi B, Henry M, El Khéchine A, Raoult D, Drancourt M (2009) High Prevalence of Methanobrevibacter smithii and Methanosphaera stadtmanae Detected in the Human Gut Using an Improved DNA Detection Protocol. PLoS ONE 4: e7063 10.1371/journal.pone.0007063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lurie-Weinberger MN, Gophna U (2015) Archaea in and on the Human Body: Health Implications and Future Directions. PLOS Pathog 11: e1004833 10.1371/journal.ppat.1004833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chandel DS, Braileanu GT, Chen JH, Chen HH, Panigrahi P (2011) Live colonocytes in newborn stool: surrogates for evaluation of gut physiology and disease pathogenesis. Pediatr Res 70: 153–158. 10.1038/pr.2011.378 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fonti R, Latella G, Bises G, Magliocca F, Nobili F, et al. (1994) Human colonocytes in primary culture: a model to study epithelial growth, metabolism and differentiation. International Journal of Colorectal Disease 9: 13–22. [DOI] [PubMed] [Google Scholar]
- 23.Nair PP (2002) Isolated colonocytes. United States Patent US6335193.
- 24.Chehoud C, Dryga A, Hwang Y, Nagy-Szakal D, Hollister EB, et al. (2016) Transfer of Viral Communities between Human Individuals during Fecal Microbiota Transplantation. mBio 7: e00322–00316. 10.1128/mBio.00322-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bihl F, Castelli D, Marincola F, Dodd RY, Brander C (2007) Transfusion-transmitted infections. Journal of Translational Medicine 5: 25–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gabriel M, Silvio DP, David H, Michael A, Moshe D, et al. (1991) Transfusion Reactions Due to Bacterial Contamination of Blood and Blood Products. Reviews of Infectious Diseases 13: 307–314. [DOI] [PubMed] [Google Scholar]
- 27.Lee CH, Steiner T, Petrof EO, et al. (2016) Frozen vs fresh fecal microbiota transplantation and clinical resolution of diarrhea in patients with recurrent Clostridium difficile infection: A randomized clinical trial. JAMA 315: 142–149. 10.1001/jama.2015.18098 [DOI] [PubMed] [Google Scholar]
- 28.Youngster I, Sauk J, Pindar C, Wilson RG, Kaplan JL, et al. (2014) Fecal Microbiota Transplant for Relapsing Clostridium difficile Infection Using a Frozen Inoculum From Unrelated Donors: A Randomized, Open-Label, Controlled Pilot Study. Clin Infect Dis 58: 1515–1522. 10.1093/cid/ciu135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sender R, Fuchs S, Milo R (2016) Are We Really Vastly Outnumbered? Revisiting the Ratio of Bacterial to Host Cells in Humans. Cell 164: 337–340. 10.1016/j.cell.2016.01.013 [DOI] [PubMed] [Google Scholar]
- 30.Ben-Amor K, Heilig H, Smidt H, Vaughan EE, Abee T, et al. (2005) Genetic diversity of viable, injured, and dead fecal bacteria assessed by fluorescence-activated cell sorting and 16S rRNA gene analysis. Appl Environ Microbiol 71: 4679–4689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satokari R, Mattila E, Kainulainen V, Arkkila PE (2015) Simple faecal preparation and efficacy of frozen inoculum in faecal microbiota transplantation for recurrent Clostridium difficile infection—an observational cohort study. Aliment Pharmacol Ther 41: 46–53. 10.1111/apt.13009 [DOI] [PubMed] [Google Scholar]
- 32.Rachmilewitz D, Katakura K, Karmeli F, Hayashi T, Reinus C, et al. (2004) Toll-like receptor 9 signaling mediates the anti-inflammatory effects of probiotics in murine experimental colitis. Gastroenterology 126: 520–528. [DOI] [PubMed] [Google Scholar]
- 33.Rachmilewitz D, Karmeli F, Takabayashi K, Hayashi T, Leider-Trejo L, et al. (2002) Immunostimulatory DNA ameliorates experimental and spontaneous murine colitis. Gastroenterology 122: 1428–1441. [DOI] [PubMed] [Google Scholar]
- 34.Gough E, Shaikh H, Manges AR (2011) Systematic review of intestinal microbiota transplantation (fecal bacteriotherapy) for recurrent Clostridium difficile infection. Clin Infect Dis 53: 994–1002. 10.1093/cid/cir632 [DOI] [PubMed] [Google Scholar]
- 35.Blekhman R, Goodrich JK, Huang K, Sun Q, Bukowski R, et al. (2015) Host genetic variation impacts microbiome composition across human body sites. Genome Biology 16: 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Goodrich JK, Waters JL, Poole AC, Sutter JL, Koren O, et al. (2014) Human genetics shape the gut microbiome. Cell 159: 789–799. 10.1016/j.cell.2014.09.053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kassam Z, Lee CH, Yuan Y, Hunt RH (2013) Fecal Microbiota Transplantation for Clostridium difficile Infection: Systematic Review and Meta-Analysis. Am J Gastroenterol 108: 500–508. 10.1038/ajg.2013.59 [DOI] [PubMed] [Google Scholar]
- 38.Moayyedi P, Surette MG, Kim PT, Libertucci J, Wolfe M, et al. (2015) Fecal Microbiota Transplantation Induces Remission in Patients With Active Ulcerative Colitis in a Randomized Controlled Trial. Gastroenterology 149: 102–109. e106 10.1053/j.gastro.2015.04.001 [DOI] [PubMed] [Google Scholar]
- 39.Reeves AE, Koenigsknecht MJ, Bergin IL, Young VB (2012) Suppression of Clostridium difficile in the gastrointestinal tracts of germfree mice inoculated with a murine isolate from the family Lachnospiraceae. Infect Immun 80: 3786–3794. 10.1128/IAI.00647-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gerding DN, Meyer T, Lee C, Cohen SH, Murthy UK, et al. (2015) Administration of spores of nontoxigenic Clostridium difficile strain M3 for prevention of recurrent C. difficile infection: a randomized clinical trial. Jama 313: 1719–1727. 10.1001/jama.2015.3725 [DOI] [PubMed] [Google Scholar]
- 41.Kim MS, Park EJ, Roh SW, Bae JW (2011) Diversity and abundance of single-stranded DNA viruses in human feces. Appl Environ Microbiol 77: 8062–8070. 10.1128/AEM.06331-11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wigington CH, Sonderegger D, Brussaard CPD, Buchan A, Finke JF, et al. (2016) Re-examination of the relationship between marine virus and microbial cell abundances. Nature Microbiology 1: 15024. [DOI] [PubMed] [Google Scholar]
- 43.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI (2012) Going viral: next generation sequencing applied to human gut phage populations. Nat Rev Microbiol 10: 607–617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Reyes A, Haynes M, Hanson N, Angly FE, Heath AC, et al. (2010) Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature 466: 334–338. 10.1038/nature09199 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Norman JM, Handley SA, Baldridge MT, Droit L, Liu CY, et al. (2015) Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 160: 447–460. 10.1016/j.cell.2015.01.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Dutilh BE, Cassman N, McNair K, Sanchez SE, Silva GG, et al. (2014) A highly abundant bacteriophage discovered in the unknown sequences of human faecal metagenomes. Nat Commun 5: 4498 10.1038/ncomms5498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sarker SA, Sultana S, Reuteler G, Moine D, Descombes P, et al. (2016) Oral Phage Therapy of Acute Bacterial Diarrhea With Two Coliphage Preparations: A Randomized Trial in Children From Bangladesh. EBioMedicine 4: 124–137. 10.1016/j.ebiom.2015.12.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Meader E, Mayer MJ, Steverding D, Carding SR, Narbad A (2013) Evaluation of bacteriophage therapy to control Clostridium difficile and toxin production in an in vitro human colon model system. Anaerobe 22: 25–30. 10.1016/j.anaerobe.2013.05.001 [DOI] [PubMed] [Google Scholar]
- 49.Nale JY, Spencer J, Hargreaves KR, Buckley AM, Trzepinski P, et al. (2015) Bacteriophage Combinations Significantly Reduce Clostridium difficile Growth In Vitro and Proliferation In Vivo. Antimicrob Agents Chemother 60: 968–981. 10.1128/AAC.01774-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hargreaves KR, Clokie MRJ (2014) Clostridium difficile phages: still difficult? Frontiers in Microbiology 5(184): 10.3389/fmicb.2014.00184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.McCallin S, Alam Sarker S, Barretto C, Sultana S, Berger B, et al. (2013) Safety analysis of a Russian phage cocktail: from metagenomic analysis to oral application in healthy human subjects. Virology 443: 187–196. 10.1016/j.virol.2013.05.022 [DOI] [PubMed] [Google Scholar]
- 52.Spellberg B, Guidos R, Gilbert D, Bradley J, Boucher HW, et al. (2008) The epidemic of antibiotic-resistant infections: a call to action for the medical community from the Infectious Diseases Society of America. Clin Infect Dis 46: 155–164. 10.1086/524891 [DOI] [PubMed] [Google Scholar]
- 53.Hill DA, Hoffmann C, Abt MC, Du Y, Kobuley D, et al. (2010) Metagenomic analyses reveal antibiotic-induced temporal and spatial changes in intestinal microbiota with associated alterations in immune cell homeostasis. Mucosal Immunol 3: 148–158. 10.1038/mi.2009.132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dethlefsen L, Huse S, Sogin ML, Relman DA (2008) The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol 6: e280 10.1371/journal.pbio.0060280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Labrie SJ, Samson JE, Moineau S (2010) Bacteriophage resistance mechanisms. Nat Rev Microbiol 8: 317–327. 10.1038/nrmicro2315 [DOI] [PubMed] [Google Scholar]
- 56.Hyman P, Abedon ST (2010) Bacteriophage host range and bacterial resistance. Adv Appl Microbiol 70: 217–248. 10.1016/S0065-2164(10)70007-1 [DOI] [PubMed] [Google Scholar]
- 57.Gaci N, Borrel G, Tottey W, O’Toole PW, Brugère J (2014) Archaea and the human gut: New beginning of an old story. World J Gastroenterol 20: 16062–16078. 10.3748/wjg.v20.i43.16062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Stewart JA, Chadwick VS, Murray A (2006) Carriage, quantification, and predominance of methanogens and sulfate-reducing bacteria in faecal samples. Lett Appl Microbiol 43: 58–63. [DOI] [PubMed] [Google Scholar]
- 59.Weaver GA, Krause JA, Miller TL, Wolin MJ (1986) Incidence of methanogenic bacteria in a sigmoidoscopy population: an association of methanogenic bacteria and diverticulosis. Gut 27: 698–704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Jhangi Sushrut G R, Glanz Bonnie, Cook Sandra, Nejad Parham, Ward Doyle, Li Ning, Gerber Georg, Bry Lynn, Weiner Howard (2014) Increased Archaea Species and Changes with Therapy in Gut Microbiome of Multiple Sclerosis Subjects. Neurology 82(10): S24–001.. [Google Scholar]
- 61.Schulze J, Sonnenborn U (2009) Yeasts in the gut: from commensals to infectious agents. Dtsch Arztebl Int 106: 837–842. 10.3238/arztebl.2009.0837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH (2014) Review article: fungal microbiota and digestive diseases. Aliment Pharmacol Ther 39: 751–766. 10.1111/apt.12665 [DOI] [PubMed] [Google Scholar]
- 63.Ott SJ, Kuhbacher T, Musfeldt M, Rosenstiel P, Hellmig S, et al. (2008) Fungi and inflammatory bowel diseases: Alterations of composition and diversity. Scand J Gastroenterol 43: 831–841. 10.1080/00365520801935434 [DOI] [PubMed] [Google Scholar]
- 64.Rosenbach A, Dignard D, Pierce JV, Whiteway M, Kumamoto CA (2010) Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell 9: 1075–1086. 10.1128/EC.00034-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.White SJ, Rosenbach A, Lephart P, Nguyen D, Benjamin A, et al. (2007) Self-Regulation of Candida albicans Population Size during GI Colonization. PLoS Pathog 3: e184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Albac S, Schmitz A, Lopez-Alayon C, d'Enfert C, Sautour M, et al. (2016) Candida albicans is able to use M cells as a portal of entry across the intestinal barrier in vitro. Cell Microbiol 18: 195–210. 10.1111/cmi.12495 [DOI] [PubMed] [Google Scholar]
- 67.Ksiadzyna D, Semianow-Wejchert J, Nawrot U, Wlodarczyk K, Paradowski L (2009) Serum concentration of interleukin 10, anti-mannan Candida antibodies and the fungal colonization of the gastrointestinal tract in patients with ulcerative colitis. Adv Med Sci 54: 170–176. 10.2478/v10039-009-0023-6 [DOI] [PubMed] [Google Scholar]
- 68.Standaert-Vitse A, Sendid B, Joossens M, Francois N, Vandewalle-El Khoury P, et al. (2009) Candida albicans colonization and ASCA in familial Crohn's disease. Am J Gastroenterol 104: 1745–1753. 10.1038/ajg.2009.225 [DOI] [PubMed] [Google Scholar]
- 69.Nair P, Lagerholm S, Dutta S, Shami S, Davis K, et al. (2003) Coprocytobiology: on the nature of cellular elements from stools in the pathophysiology of colonic disease. J Clin Gastroenterol 36: S84–93; discussion S94-86. [DOI] [PubMed] [Google Scholar]
- 70.Maynard CL (2012) Reciprocal Interactions of the Intestinal Microbiota and Immune System. 489: 231–241. 10.1038/nature11551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Magnusson M, Magnusson KE, Sundqvist T, Denneberg T (1991) Impaired intestinal barrier function measured by differently sized polyethylene glycols in patients with chronic renal failure. Gut 32: 754–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Vaziri ND, Zhao YY, Pahl MV (2015) Altered intestinal microbial flora and impaired epithelial barrier structure and function in CKD: the nature, mechanisms, consequences and potential treatment. Nephrol Dial Transplant 31: 737–746. 10.1093/ndt/gfv095 [DOI] [PubMed] [Google Scholar]
- 73.Yui S, Nakamura T, Sato T, Nemoto Y, Mizutani T, et al. (2012) Functional engraftment of colon epithelium expanded in vitro from a single adult Lgr5(+) stem cell. Nat Med 18: 618–623. 10.1038/nm.2695 [DOI] [PubMed] [Google Scholar]
- 74.Jung P, Sato T, Merlos-Suarez A, Barriga FM, Iglesias M, et al. (2011) Isolation and in vitro expansion of human colonic stem cells. Nat Med 17: 1225–1227. 10.1038/nm.2470 [DOI] [PubMed] [Google Scholar]
- 75.Mantis NJ, Rol N, Corthésy B (2011) Secretory IgA's Complex Roles in Immunity and Mucosal Homeostasis in the Gut. Mucosal immunology 4: 603–611. 10.1038/mi.2011.41 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Dallas SD, Rolfe RD (1998) Binding of Clostridium difficile toxin A to human milk secretory component. Journal of Medical Microbiology 47: 879–888. [DOI] [PubMed] [Google Scholar]
- 77.Olson A, Diebel LN, Liberati DM (2013) Effect of host defenses on Clostridium difficile toxin-induced intestinal barrier injury. J Trauma Acute Care Surg 74: 983–989; discussion 989–990. 10.1097/TA.0b013e3182858477 [DOI] [PubMed] [Google Scholar]
- 78.Maslowski KM, Vieira AT, Ng A, Kranich J, Sierro F, et al. (2009) Regulation of inflammatory responses by gut microbiota and chemoattractant receptor GPR43. Nature 461: 1282–1286. 10.1038/nature08530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Arpaia N, Campbell C, Fan X, Dikiy S, van der Veeken J, et al. (2013) Metabolites produced by commensal bacteria promote peripheral regulatory T-cell generation. Nature 504: 451–455. 10.1038/nature12726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Furusawa Y, Obata Y, Fukuda S, Endo TA, Nakato G, et al. (2013) Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 504: 446–450. 10.1038/nature12721 [DOI] [PubMed] [Google Scholar]
- 81.Sonnenburg ED, Smits SA, Tikhonov M, Higginbottom SK, Wingreen NS, et al. (2016) Diet-induced extinctions in the gut microbiota compound over generations. Nature 529: 212–215. 10.1038/nature16504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Sonnenburg ED, Sonnenburg JL (2014) Starving our microbial self: the deleterious consequences of a diet deficient in microbiota-accessible carbohydrates. Cell Metab 20: 779–786. 10.1016/j.cmet.2014.07.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Horner KL, LeBoeuf RC, McFarland LV, Elmer GW (2000) Dietary fiber affects the onset of Clostridium difficile disease in hamsters. Nutrition Research 20: 1103–1112. [Google Scholar]
- 84.Nicolucci AC, Reimer RA (2016) Prebiotics as a modulator of gut microbiota in paediatric obesity. Pediatr Obes 10.1111/ijpo.12140 [DOI] [PubMed] [Google Scholar]
- 85.Nicolucci AC, Hume MP, Reimer RA (2015) Effect of Prebiotic Fiber Intake on Adiposity and Inflammation in Overweight and Obese Children: Assessing the Role of the Gut Microbiota. Canadian Journal of Diabetes 39: S43. [Google Scholar]
- 86.Machiels K, Joossens M, Sabino J, De Preter V, Arijs I, et al. (2014) A decrease of the butyrate-producing species Roseburia hominis and Faecalibacterium prausnitzii defines dysbiosis in patients with ulcerative colitis. Gut 63: 1275–1283. 10.1136/gutjnl-2013-304833 [DOI] [PubMed] [Google Scholar]
- 87.Sokol H, Seksik P, Furet JP, Firmesse O, Nion-Larmurier I, et al. (2009) Low counts of Faecalibacterium prausnitzii in colitis microbiota. Inflamm Bowel Dis 15: 1183–1189. 10.1002/ibd.20903 [DOI] [PubMed] [Google Scholar]
- 88.Weir TL, Manter DK, Sheflin AM, Barnett BA, Heuberger AL, et al. (2013) Stool microbiome and metabolome differences between colorectal cancer patients and healthy adults. PLoS ONE 8: e70803 10.1371/journal.pone.0070803 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Vernia P, Marcheggiano A, Caprilli R, Frieri G, Corrao G, et al. (1995) Short-chain fatty acid topical treatment in distal ulcerative colitis. Aliment Pharmacol Ther 9: 309–313. [DOI] [PubMed] [Google Scholar]
- 90.Scheppach W, Sommer H, Kirchner T, Paganelli GM, Bartram P, et al. (1992) Effect of butyrate enemas on the colonic mucosa in distal ulcerative colitis. Gastroenterology 103: 51–56. [DOI] [PubMed] [Google Scholar]
- 91.Harig JM, Soergel KH, Komorowski RA, Wood CM (1989) Treatment of diversion colitis with short-chain-fatty acid irrigation. N Engl J Med 320: 23–28. [DOI] [PubMed] [Google Scholar]
- 92.Breuer RI, Soergel KH, Lashner BA, Christ ML, Hanauer SB, et al. (1997) Short chain fatty acid rectal irrigation for left-sided ulcerative colitis: a randomised, placebo controlled trial. Gut 40: 485–491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Patz J, Jacobsohn WZ, Gottschalk-Sabag S, Zeides S, Braverman DZ (1996) Treatment of refractory distal ulcerative colitis with short chain fatty acid enemas. Am J Gastroenterol 91: 731–734. [PubMed] [Google Scholar]
- 94.Vieira EL, Leonel AJ, Sad AP, Beltrao NR, Costa TF, et al. (2012) Oral administration of sodium butyrate attenuates inflammation and mucosal lesion in experimental acute ulcerative colitis. J Nutr Biochem 23: 430–436. 10.1016/j.jnutbio.2011.01.007 [DOI] [PubMed] [Google Scholar]
- 95.Haghikia A, Jorg S, Duscha A, Berg J, Manzel A, et al. (2015) Dietary Fatty Acids Directly Impact Central Nervous System Autoimmunity via the Small Intestine. Immunity 43: 817–829. 10.1016/j.immuni.2015.09.007 [DOI] [PubMed] [Google Scholar]
- 96.Fröhlich EE, Mayerhofer R, Holzer P (2015) Reevaluating the hype: four bacterial metabolites under scrutiny. European Journal of Microbiology & Immunology 5: 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Tremaroli V, Backhed F (2012) Functional interactions between the gut microbiota and host metabolism. Nature 489: 242–249. 10.1038/nature11552 [DOI] [PubMed] [Google Scholar]
- 98.Lupton JR (2004) Microbial degradation products influence colon cancer risk: the butyrate controversy. J Nutr 134: 479–482. [DOI] [PubMed] [Google Scholar]
- 99.Donia MS, Fischbach MA (2015) Small Molecules from the Human Microbiota. Science 349(6246): 1254766(1–10). 10.1126/science.1254766 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Ridaura VK, Faith JJ, Rey FE, Cheng J, Duncan AE, et al. (2013) Gut microbiota from twins discordant for obesity modulate metabolism in mice. Science 341(6150): 10.1126/science.1241214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Kohl KD, Stengel A, Dearing MD (2015) Inoculation of tannin-degrading bacteria into novel hosts increases performance on tannin-rich diets. Environmental Microbiology 10.1111/1462-2920.12841 [DOI] [PubMed] [Google Scholar]
- 102.den Besten G, van Eunen K, Groen AK, Venema K, Reijngoud DJ, et al. (2013) The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J Lipid Res 54: 2325–2340. 10.1194/jlr.R036012 [DOI] [PMC free article] [PubMed] [Google Scholar]