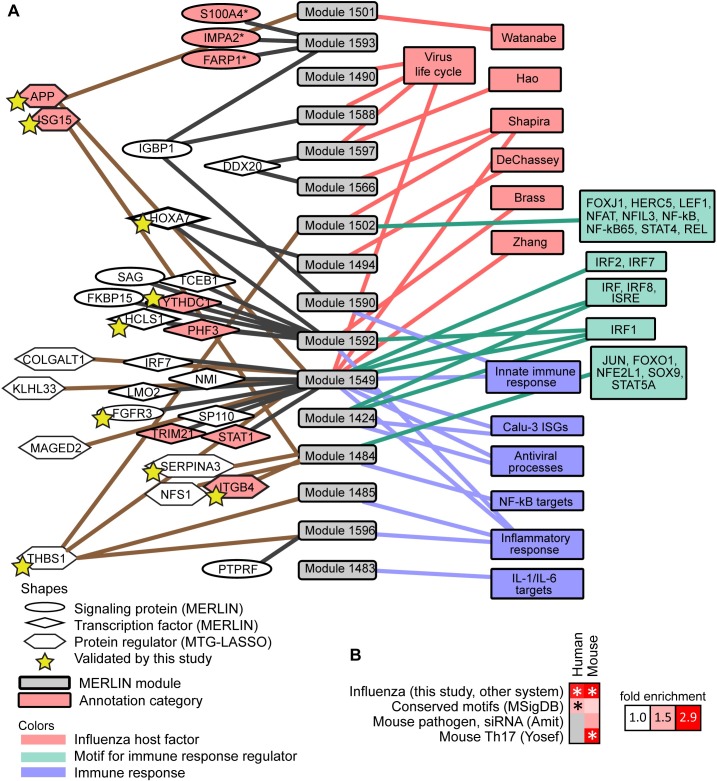
Fig 3. Overview of human influenza response modules and top predicted regulators.
A. A selection of sixteen human modules (grey, center), with their top predicted regulators (shapes on left, connected to modules), and enriched gene sets relevant to immune response or viral life cycle (colored boxes, right). Modules were included in this figure if they were enriched for one of the following categories: (a) influenza host genes (pink) identified by RNAi, protein-protein interactions, expert curation (Zhang et al. 2009), or enrichment for other virus life cycle gene sets (from GO or KEGG); (b) motifs (green) for transcriptional regulators whose annotations indicate relevance to the innate immune response; (c) innate immune response gene sets (blue) from experimental results or manual curation. One module, Cluster 1593, is also included because its consensus regulator set (but not targets) was enriched for host proteins that are involved in the influenza life cycle from Watanabe et al (2014). Also shown are seventeen (ellipses and diamonds) predicted mRNA-level regulators, and ten predicted protein regulators (hexagons), that are associated with these modules. Regulators with evidence for influenza relevance are shaded in pink. Host genes that significantly impact viral replication identified by this study (APP, FGFR3, HCLS1, HOXA7, SERPINA3) are indicated with yellow stars. B. Comparison of the MERLIN inferred mouse and human influenza response networks ("Influenza (this study, other system)") to each other and to other regulatory networks. Fold enrichment of shared edges over expected fraction is visualized in the heat map, with significant comparisons marked with asterisks (hypergeometric p-value < 0.05).