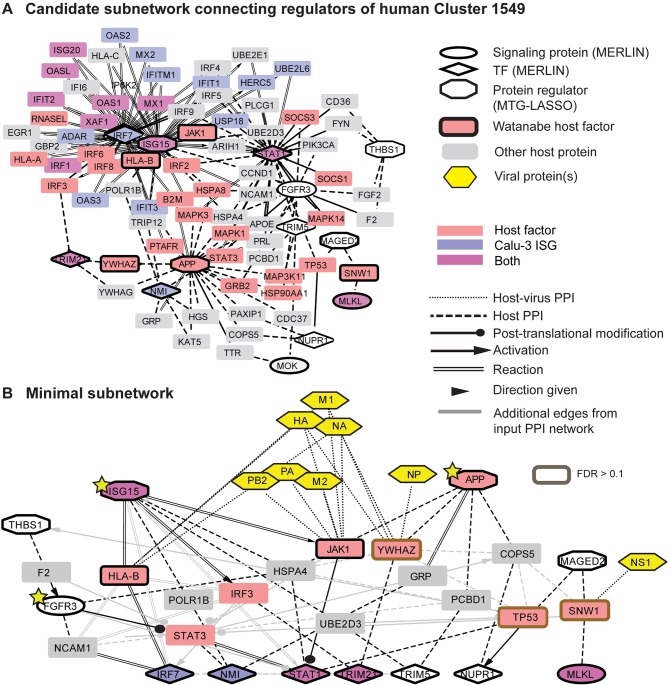
Fig 7. Integration of expression- and protein-based regulators into a physical subnetwork for module 1549.
A. Candidate subnetwork for module 1549 given as input to the Integer Linear Programming (ILP) approach to find a minimal network. The candidate subnetwork is extracted from the background interaction network by connecting MERLIN (mRNA) and MTG-LASSO (protein) regulators through at most one intermediate node and mRNA signaling proteins to TFs (Materials and Methods). Nodes with dark outlines are input to the ILP-based subnetwork inference method: MERLIN regulators (diamonds and ellipses), protein regulators (octagons), and host genes for influenza identified by Watanabe et al (2014) (boxes). Nodes without borders are candidate intermediates between two regulators and were not given special treatment by the method. Node color indicates host genes involved in influenza obtained from siRNA or protein interaction studies and Calu-3 ISGs (as in Fig 3); this information is for visualization purposes and is not used in the subnetwork inference method. Only edges between intermediate nodes and input nodes are shown. Additional available protein-protein interactions between intermediate nodes are not shown for visual clarity. B. Minimal subnetwork for Cluster 1549's regulators inferred using ILP. Yellow stars indicate host genes for which siRNA knockdown significantly impacted viral replication. Also shown are interactions between Watanabe host genes and viral proteins (yellow hexagons). Grey edges are input to the method but not selected by the approach. All selected edges and all nodes other than YWHAZ, TP53, SNW1 (brown boxes) had FDR ≤ 0.10. FDR was assessed based on permutation tests.