Figure 2. Quantifying Pol II transcription kinetics in vivo.
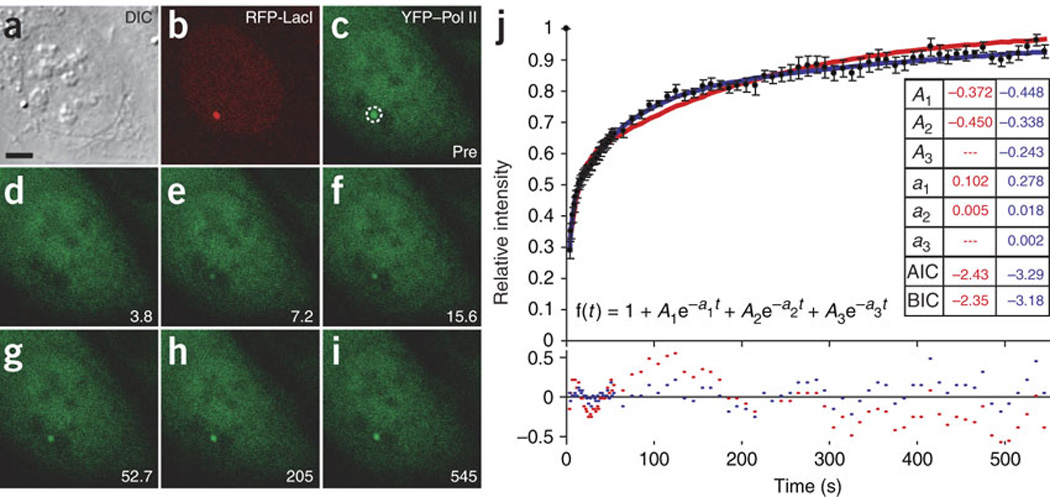
Fluorescence recovery after photobleaching of the transcription site is shown in a–i. (a) Differential interference contrast images of live cells. (b) RFP-LacI labels gene locus. (c) Dashed circle indicates photobleached region. (d–i) Bleaching (d) and recovery (e–i) of YFP–Pol II17 at active site, monitored for 545 s. Scale bar, 5 µm. (j) Pol II FRAP data (black; n = 10) fit to a sum of exponentials (see equation) to determine the minimal model complexity. This was done using generalized least-squares optimization as implemented in the SAAM II software package (http://depts.washington.edu/saam2/). Goodness of fit was evaluated by requiring that coefficients of variation on the parameter estimates were less than 30% and by checking for a random distribution of residuals around 0 (red and blue dots in lower chart represent residuals for two and three exponentials, respectively). By these criteria, a fit of the Pol II FRAP data requires three exponentials (blue), as residuals are not randomly distributed when fit to two exponentials (red). The Akaike information criterion (AIC)59 and the Bayes-Schwarz information criterion (BIC)60 for two- and three-exponential models are reported in the inset table. These standard quantitative measures of goodness-of-fit penalize additional model parameters. If the fit is sufficiently improved to justify the increased complexity of the model, then the AIC and BIC of the more complex model will be less than those of the simpler model. By this measure, three exponentials are superior to two in modeling our data. Error bars show s.e.m.
