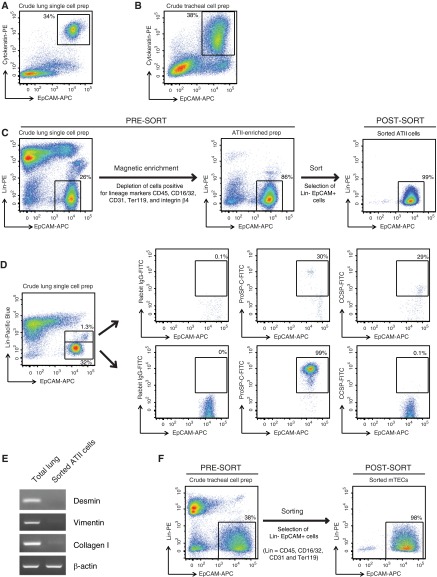
Figure 1.
Cell-sorting strategy for isolating highly pure mouse alveolar type (AT) II cells and murine tracheal epithelial cells (mTECs). Representative flow cytometric dot plots of (A) a crude single-cell preparation of mouse lung cells or (B) tracheal cell preparations, stained on the surface for epithelial cell adhesion molecule (EpCAM)-allophycocyanin (APC) and intracellularly for cytokeratin-phycoerythrin (PE). (C) ATII cell–sorting strategy. Shown are representative dot plots obtained from flow cytometric analysis of lung cells collected at various stages of ATII cell isolation (pre- and post-magnetic enrichment, and post-sort) and stained for lineage (Lin) markers (CD45, CD16/32, CD31, Ter119, and integrin β4) versus EpCAM. Events displayed on the plots were 4′,6-diamidino-2-phenylindole–negative single cells. (D) Exclusion of club cell contamination during fluorescence-activated cell sorting for ATII cells. Representative flow cytometric dot plots of a crude single-cell preparation of lung cells stained for lineage markers (CD45, CD16/32, CD31, Ter119, and integrin β4) in the pacific blue channel and EpCAM-APC. The cells were also stained intracellularly for either rabbit isotype IgG, pro-surfactant protein (SP)-C or club cell secretory protein (CCSP), in the fluorescein isothiocyanate (FITC) channel and the distribution of this staining is shown in both the lower Lin− EpCAM+ gate, which was used to sort ATII cells, and the upper Linlow EpCAM+ gate. (E) RT-PCR analysis for the indicated genes was conducted on the highly pure, flow-sorted ATII cells, as described in Materials and Methods. Data shown are representative of three independent experiments. (F) Shown are flow cytometric dot plots from a representative cell sort performed for the isolation of highly pure mTECs.