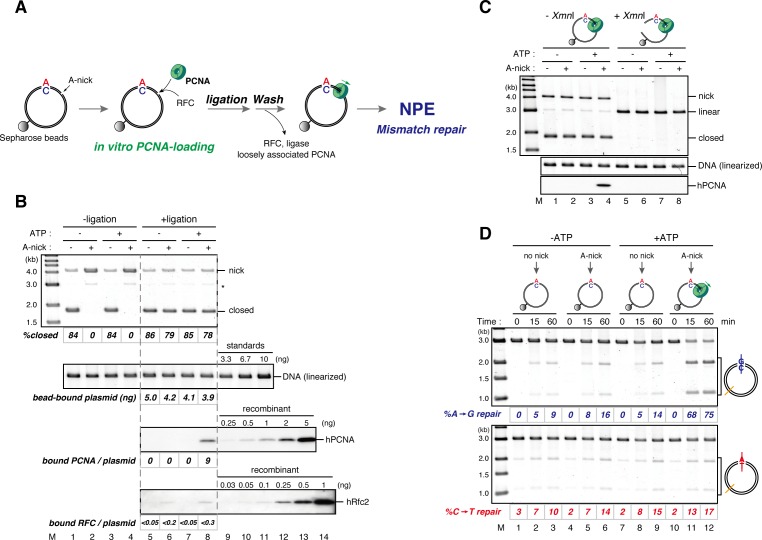
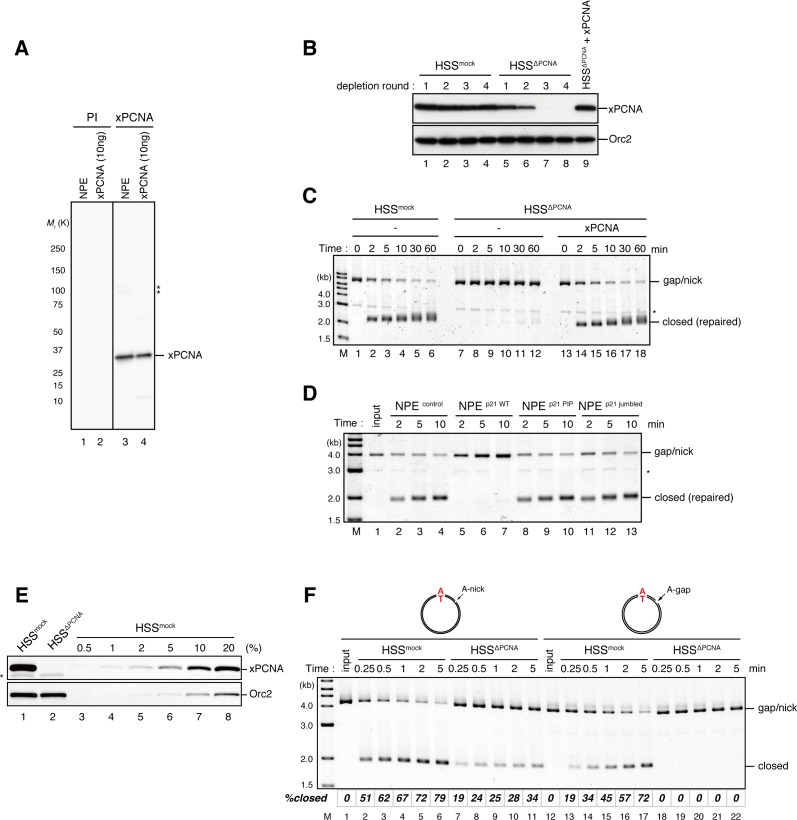
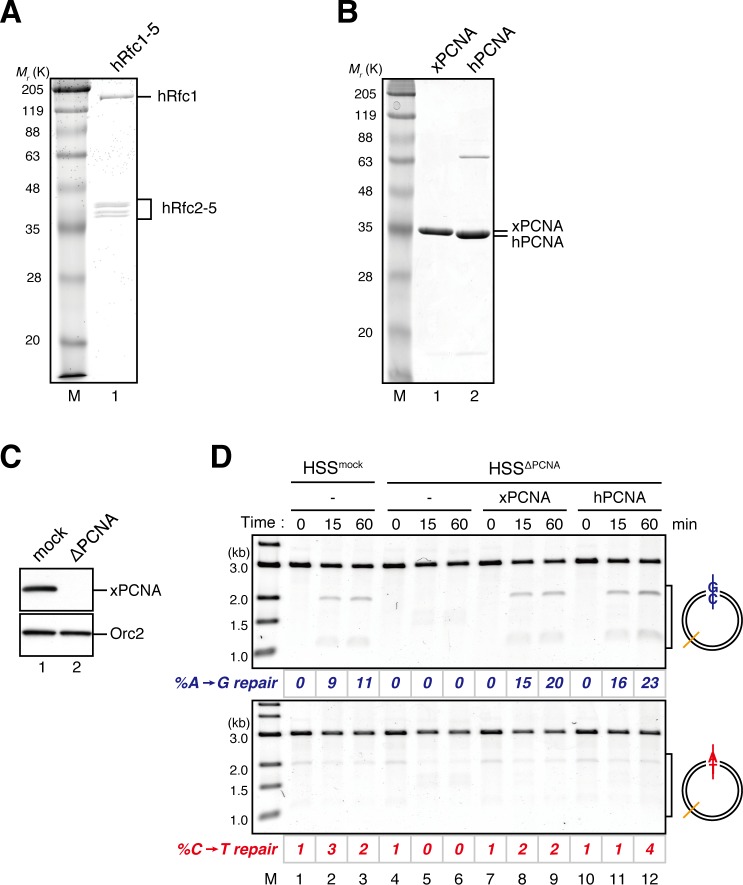
(
A) Characterization of xPCNA sera. 0.1 μl of NPE and 10 ng of recombinant xPCNA were separated using 4–15% SDS-PAGE and transferred onto PVDF membranes. Each membrane strip was probed with either the xPCNA or pre-immune serum (PI) from the same rabbit. The same exposure sets of PI and xPCNA antibodies are shown. xPCNA (
Mr = 2.88 × 10
4 as a monomer) was detected as nearly a single band in NPE. (*) indicates cross-reacting band. (
B) The efficiency of PCNA depletion from HSS. HSS was depleted with pre-immune (lanes 1–4) or xPCNA antibodies (lanes 5–9), and supplemented with either control buffer (lanes 4 and 8) or 0.5 μM recombinant xPCNA (lane 9). HSS (0.5 μl) from each depletion rounds was separated by SDS-PAGE and probed with the indicated antibodies. Orc2 served as a loading control. (
C) The gap-filling reaction in PCNA-depleted HSS. Gap-carrying homoduplex pMM1
AT plasmids were incubated in PCNA-depleted HSS described in (
B) and sampled at the indicated times. The efficiency of the gap-filling reaction was analyzed using ethidium bromide containing agarose gel electrophoresis. Depletion of PCNA inhibited appearance of closed-form plasmids even after 60 min (lane 12), indicating that PCNA is essential for either DNA synthesis at the gap site, flap-processing, or ligation at the gap in HSS. (*) indicates linearized fragments produced by the nicking endonuclease. (
D) The gap-filling reaction in p21-peptide supplemented NPE. Because PCNA-depletion from NPE was not possible due to its high concentration, p21 peptides (
Mattock et al. 2001) were used to inhibit PCNA function in NPE. Gap-carrying homoduplex pMM1
AT plasmids were incubated in NPE supplemented with either control buffer (lanes 2–4), 1 mg/ml wildtype (lanes 5–7), PIP mutant (lanes 8–10) or jumbled p21 peptide (lanes 11–13), and sampled at the indicated times. The efficiency of gap filling was analyzed using ethidium bromide containing agarose gel electrophoresis. The wildtype but not the PIP mutant nor jumbled p21 peptide inhibited appearance of closed-form plasmids, suggesting that PCNA is required for either DNA synthesis at the gap site, flap-processing, or ligation at the gap in NPE. (*) indicates linearized fragments produced by the nicking endonuclease. (
E) The depletion efficiency of PCNA from HSS. HSS was depleted using pre-immune (lane 1) or xPCNA antibodies (lane 2). Immunoblots of the HSS samples (0.5 μl each) are shown. Orc2 served as a loading control. (*) indicates cross-reacting band. (
F) Comparison of nick ligation and gap filling in PCNA-depleted HSS. Nick-carrying homoduplex pMM2
AT (lanes 1–11) or gap-carrying homoduplex pMM1
AT (lanes 12–22) was incubated in either mock-treated or PCNA-depleted HSS described in (
E), and sampled at the indicated times. The efficiencies of accumulation of closed-circular molecules were analyzed using ethidium bromide containing agarose gel electrophoresis. Although gap filling was completely inhibited by PCNA-depletion, nick ligation was only partially inhibited, indicating that PCNA is only partially required for nick ligation in HSS.