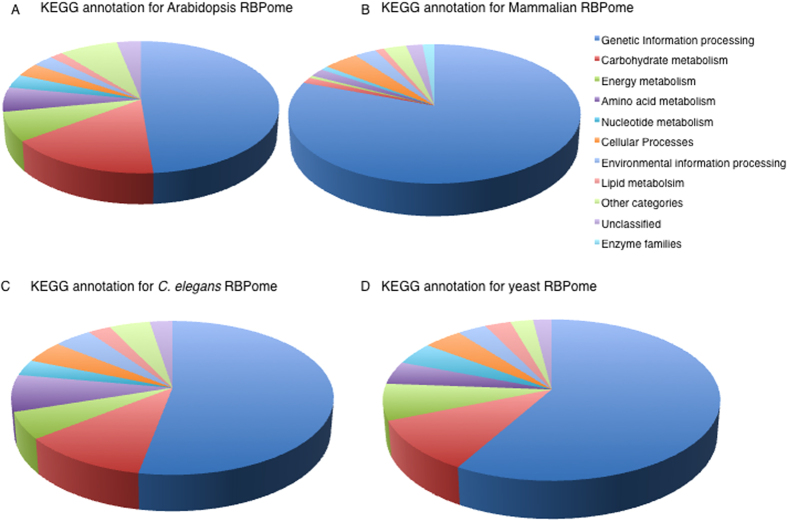
Figure 4. Kyoto encyclopedia of genes and genomes (KEGG) pathway analysis.
(A) KEGG annotation for Arabidopsis RBP representing 58% of identified proteins that were successfully assigned to pathways. (B) KEGG annotation for mammalian RBP representing 78% of identified proteins that were successfully assigned to pathways. (C) KEGG annotation for C. elegans RBP representing 85% of the identified proteins that could be assigned to pathways. (D) KEGG annotation for yeast RBP representing 79% of the identified proteins that were assigned to pathways. The mapped set of proteins include intermediary metabolism pathway and the analysis was performed using KEGG mapper (http://www.kegg.jp/).