Abstract
Previous mass spectrometry analyses described human mitochondrial peptides entirely translated from swinger RNAs, RNAs where polymerization systematically exchanged nucleotides. Exchanges follow one among 23 bijective transformation rules, nine symmetric exchanges (X ↔ Y, e.g. A ↔ C) and fourteen asymmetric exchanges (X → Y → Z → X, e.g. A → C → G → A), multiplying by 24 DNA's protein coding potential. Abrupt switches from regular to swinger polymerization produce chimeric RNAs. Here, human mitochondrial proteomic analyses assuming abrupt switches between regular and swinger transcriptions, detect chimeric peptides, encoded by part regular, part swinger RNA. Contiguous regular- and swinger-encoded residues within single peptides are stronger evidence for translation of swinger RNA than previously detected, entirely swinger-encoded peptides: regular parts are positive controls matched with contiguous swinger parts, increasing confidence in results. Chimeric peptides are 200 × rarer than swinger peptides (3/100,000 versus 6/1000). Among 186 peptides with > 8 residues for each regular and swinger parts, regular parts of eleven chimeric peptides correspond to six among the thirteen recognized, mitochondrial protein-coding genes. Chimeric peptides matching partly regular proteins are rarer and less expressed than chimeric peptides matching non-coding sequences, suggesting targeted degradation of misfolded proteins. Present results strengthen hypotheses that the short mitogenome encodes far more proteins than hitherto assumed. Entirely swinger-encoded proteins could exist.
Keywords: RNA–DNA differences, Bijective transformation, Nucleotide substitution, Proteome, Systematic deletions, delRNA
Graphical abstract
Highlights
-
•
Chimeric peptides are translated from contiguous regular and swinger RNA
-
•
They are 200x rarer than mitochondrial swinger peptides
-
•
Chimeric peptides integrated in regular mitochondrial proteins are downregulated
-
•
Contiguous regular parts are matched positive controls for swinger parts
-
•
The last point validates results beyond other statistical tests for robustness
1. Introduction
Mitochondrial genomes apparently compensate for their reduced size by cumulating multiple functions for single sequences [12], [14]. For example, tDNA, DNA templating for tRNAs, probably functions also occasionally as light strand replication origin [79], [80], [83], [106], [107], [108], [110]. The complementary strand of tDNA has similar secondary structure formation capacities and might template for additional functional tRNAs with anticodons usually corresponding to the inverse complement of the tRNA's regular anticodon [7], [81], [82], [84]. Mitochondrial tRNA sidearm loops might also function as anticodons, potentially increasing further mitochondrial anticodon repertoires [90], [95]. Translation of stop codons also increases protein coding repertoires [81], reassigning stop codons to amino acids [26], [84], [85], [86], [87], [91], [7], [11], [98].
These various mechanisms expand protein-coding potentials of DNA/RNA sequences. Multifunctional sequences, as suggested for tRNA synthetase genes [53], [69], [70] are presumably relicts of ancient, short protogenomes, plausibly consisting of ancestors of ribosomal RNAs [72], [73], [111], where sequence multifunctionality was probably essential. Presumably, alternative codings are relicts of mechanisms that increase sequence multifunctionality.
1.1. Swinger polymerization
A further little known phenomenon increases DNA's protein coding repertoire: nucleotide polymerization that systematically exchanges nucleotides. This alters gene and mRNA coding properties. Assuming this phenomenon enables to detect homology relationships of otherwise ‘orphan’ DNA and RNA sequences. The homology of these orphan sequences had not been determined because these apparently orphan sequences are so much transformed as compared to their ‘parent’ homologue that homology is undetectable without assuming a systematic exchange between nucleotides, but becomes obvious after taking the systematic exchange(s) into account. These transformations consist of systematic exchanges between nucleotides during DNA or RNA polymerization, producing so-called swinger sequences.
The first described swinger RNAs were from vertebrate mitogenomes, and correspond to a 3′-to -5′ inversion, without complementing, of the homologous, template sequence [88], [92], also called ‘reversing’ transformation [27], [28]. When considering a specific sequence, this transformation follows the swinger rule A ↔ T + C ↔ G (bijective transformation rule π9 according to the annotation system in [58]) of the negative strand of the specific sequence, one among nine systematic symmetric exchanges, of type X ↔ Y, e.g. A ↔ C [93]. Fourteen asymmetric exchanges exist, of type X → Y → Z → X, e.g. A → C → G > A [94]. About hundred mitochondrial transcripts corresponding to one of these 23 swinger types have been detected within the human EST database of GenBank, with about twice as many from the nine symmetric exchanges than from the fourteen asymmetric exchanges.
Swinger RNAs matching eleven exchange types were detected within GenBank's EST database, six symmetric, and four asymmetric transformations. Most of these swinger RNAs (obtained by classical Sanger sequencing) are longer than 100 nucleotides and have > 90% similarity with the mitogenome if the swinger transformation is assumed over their complete length [90], [91]. All 23 swinger types exist in the human mitochondrial transcriptome among short reads produced by RNA seq (Illumina) ([103], data from [30]). Abundances of different swinger types as estimated from GenBank's ESTs (sequenced by classical methods) and next generation massive sequencing (RNA seq) are overall congruent (i.e. Figure 2 in [103]). This congruence between swinger RNA abundances is remarkable for two reasons: first because comparable results are obtained by two independent methods (Sanger versus next generation sequencing); and second because biological samples differ (not the same cells/mitochondrial lines were analyzed). This suggests that mitochondrial swinger transcription is general to mitochondria, not tissue- or line-specific.
Hence sequences potentially template for 23 swinger transformed versions, increasing considerably the potential coding density of any sequence. Swinger DNA was also detected for nuclear and mitochondrial genes [96], [97], especially ribosomal RNAs [99], but for now only according to swinger rule A ↔ T + C ↔ G. Swinger sequences detected in Genbank originate from numerous independent research projects and laboratories, only this author describes them as swinger-transformed.
1.2. Swinger versus chimeric RNAs and peptides
Some detected sequences are not entirely swinger-transformed, sequences contiguous to the swinger sequence match the untransformed, contiguous DNA template, and hence are regular RNA [100]. These RNAs transcribed partly by regular, and partly by swinger transcriptions, are termed chimeric RNAs [100]. The transition from one to the other part is frequently abrupt, suggesting sudden switches in the polymerization mode of the same polymerase.
Analyzes here search for peptides matching translation of such chimeric RNAs, where contiguous parts of the peptide are translated from regular and swinger parts of the sequence. These peptides are also considered chimeric, and differ from previously described swinger peptides [103] because the latter are only translated from swinger-transformed RNA, while chimeric peptides would be transcribed from RNA that is in part regular, and in part swinger-transformed.
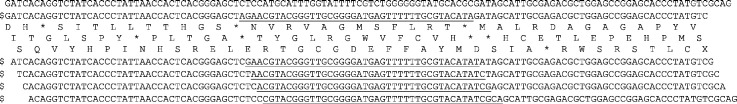
The principle according to which chimeric sequences are produced is shown for a specific 120 nucleotides long sequence of the human mitogenome (Fig. 1). The mid-forty nucleotides are swinger transformed according to swinger rule A ↔ C + G ↔ T (second sequence in Fig. 1). Swinger RNAs consist solely of swinger-transformed regions such as the underlined transformed regions. Chimeric RNAs have at least one of the contiguous, untransformed (5′ and/or 3′) parts. Swinger peptides are solely translated from swinger-transformed sequences (such as the underlined sequence in Fig. 1). Chimeric peptides are translated from a sequence that stretches over a regular and a swinger-transformed RNA region. Only a minority of detected RNA sequences bearing swinger transformations are chimeric, most follow in their entirety a given nucleotide exchange rule [90], [91], [100]. Detection of chimeric peptides would be evidence independent of previous descriptions of swinger peptides (entirely encoded by swinger-transformed RNA [103]) for translation of swinger-transformed RNA. Much fewer chimeric human mitochondrial RNAs [100] than entirely swinger-transformed human mitochondrial RNAs [90], [91] have been detected. Hence I expect to detect fewer chimeric peptides than for previous analyzes searching for entirely swinger-encoded peptides.
Fig. 1.
Example of running windows reduced to 120 nucleotides for illustration purposes, for the human mitochondrial genome, and swinger transformation of the mid-third 40 nucleotides. First row: regular genomic sequence; rows starting by $ are the first 5 running windows, with the mid third swinger transformed according to swinger rule A ↔ C + G ↔ T (as an example). Running windows used for actual analyses are 270 nucleotides long, and transformed according to each of the 23 swinger transformations, along the same principles as shown above for 120 nucleotides. The three peptides translated from the first running window sequence are also indicated, stops codons are translated here as ‘*’. Analyses searching for mass spectrometry data matching these predicted peptides consider the possibility that any amino acid is integrated at stops (19 possibilities, merging leucine and isoleucine, which are undistinguishable by the sequencing technique used here, because their molecular weights are identical). This means that for a single running window sequence after a single swinger transformation, 3 × 19 = 57 hypothetical peptides are considered. This number is doubled to 114 considering the inverse complement sequence, and multiplied by 23 considering all 23 potential swinger transformations. Hence 2622 peptides are translated from the 23 swinger transformations of each running window sequence indicated by $. This running window structure enables detection of chimeric peptides where the regular part is translated from the 5′, as well as from the 3′ side of the swinger part.
1.3. Swinger polymerization by regular polymerases?
Swinger polymerizations could result from unusual polymerization modes by regular polymerases because the principle of swinger polymerization does not differ from that of point nucleotide misinsertions. The difference is in the systematic change in templating rules, from f(A,C,G,T) = (A,C,G,T/U) (regular DNA replication/transcription), to a different rule, e.g. A ↔ C, which can also be annotated as f(A,C,G,T/U) = (C,A,G,T/U), stressing its systematic, rather than punctual nature. It seems plausible that point nucleotide misinsertions are due to switches to unstable, unusual conformations of polymerases, lasting the time of a misinsertion. Hypothetically, these unstable, misinsertion-inducing conformations are occasionally stabilized, so that the nucleotide exchange corresponding to that misinsertion occurs systematically along the sequence stretch polymerized while the polymerase is in that unusual conformation, producing a swinger DNA/RNA. Swinger RNAs are for now the only evidence indicating the existence of such unusual, stabilized polymerase states.
This hypothesis on polymerase conformations yields two testable predictions. The first prediction is that biochemical parameters experimentally estimated for point misinsertions by polymerases predict properties of swinger sequences. In this respect, the affinity (Km) and Vmax of each of the twelve misinsertions, and the four regular insertions, as determined by Lee and Johnson [46]) (therein Table 1), were used to predict abundances of swinger RNAs. Indeed, these experimental kinetic parameters predict several properties of swinger RNAs [58], [93], [94], strengthening the hypothesis that regular polymerases are responsible for swinger polymerizations, by switching to unusual, stabilized ‘swinger’ conformations, similar (or even identical) to conformations causing point misinsertions, but lasting longer.
Table 1.
Human mitochondrial peptides detected assuming abrupt switches between regular and swinger parts of RNA, for peptides where each regular and swinger parts have > 8 amino acids (mass spectra from [33]). Columns are: 1. Peptide number; 2. swinger type; 3. amino acid inserted at stop(s) (‘no’ indicates lack of stops); 4. strand and frame; 5. peptide sequence; 6. PSMs; 7. Xcorr; 8. trypsin miscleavage; 9. PEP; 10–13. Positive strand positions of 5’ and 3’ extreme amino acids of regular and swinger parts of detected peptide; 14. Peptide extremity matching regular transcription. Underlined: peptide swinger part; *, $ marks swinger peptide parts covering previously described swinger reads, respectively previously described swinger peptides [103]. Peptides 8 and 9 differ in posttranslational amino acid modifications (not indicated). Highlighted peptide parts match both translations according to vertebrate mitochondrial, and nuclear (standard) genetic codes. Peptide parts not highlighted match only translation according to the vertebrate mitochondrial genetic code, and are incompatible with translation according to the nuclear genetic code. For example, peptide 3 could be translated in the cytosol on the base of RNA transcribed from mitochondrial inserts in the nuclear chromosome (numts), peptides 5 could not, as peptides 1 and 2 because at least one part of the peptide is not compatible with translation according to the nuclear genetic code. Further analyses (see text) show that fewer detected peptides are compatible with the nuclear genetic code than expected by chance, and that more peptides than expected by chance are compatible only with translation according to the mitochondrial genetic code.
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | 13 | 14 | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | ac | a | r0 | YGVSEGLAAPVGAYNVGAFAALYMAANFSFLNQDAVQVADK | 3 | 2.77 | 0 | 0.374 | 3246 | 3156 | 3153 | 3123 | 5′ | 29 | 12 |
| 2 | ac | v | r1 | SCLLAFLMGSLMLTLIVGLSK | 20 | 3.04 | 0 | 0.169 | 3465 | 3432 | 3492 | 3468 | 3′ | 12 | 9 |
| 3 | ac | r | r0 | AWGGGFDVDWWGSDDIVAMR | 43 | 3.06 | 0 | 0.745 | 16188 | 16161 | 16158 | 16131 | 5′ | 10 | 10 |
| 4 | ag | k | r2 | AIGKVAFSTSVMLEVMFLVNK | 185 | 3.06 | 1 | 0.082 | 2289 | 2262 | 2319 | 2292 | 3′ | 11 | 10 |
| 5 | ag | f | r1 | SYTFPPGSSSVACWLGCSPSPTLTLIFGLSK | 49 | 3.38 | 0 | 1.000 | 3456 | 3432 | 3522 | 3459 | 3′ | 9 | 22 |
| 6 | ag | s | r0 | GGSPSDSTTSSQQLLSSILWSK | 149 | 3.98 | 0 | 0.755 | 10125 | 10101 | 10164 | 10128 | 3′ | 10 | 12 |
| 7 | ag | no | f0 | LLGAVPLASASLTIGSLALAGMPFLTGFYSKDHIIETANMS | 1 | 2.37 | 1 | 0.931 | 13704 | 13767 | 13644 | 13701 | 3′ | 29 | 12 |
| 8 | at | no | f1 | FIAYHSPGKVNFVPATAVTR | 399 | 2.77 | 1 | 0.196 | 882 | 924 | 855 | 879 | 3′ | 11 | 9 |
| 9 | at | no | f1 | FIAYHSPGKVNFVPATAVTR | 604 | 2.77 | 1 | 0.633 | 882 | 924 | 855 | 879 | 3′ | 11 | 9 |
| 10 | at | p | f1 | MPNSFNWDVGGNSSKLPVECLVEQGPEAR | 1 | 2.31 | 1 | 0.605 | 1467 | 1500 | 1407 | 1464 | 3′ | 17 | 12 |
| 11 | at | y | r2 | TMSYALTLLLLQTCRGFSR | 25 | 4.34 | 1 | 0.836 | 5601 | 5574 | 5631 | 5604 | 3′ | 9 | 10 |
| 12 | at | no | r2 | DMGDASVMGLSVNEASYDGK | 19 | 2.17 | 0 | 0.121 | 6933 | 6906 | 6963 | 6936 | 3′ | 10 | 10 |
| 13 | cg | no | f0 | TLGQGVAHDLRTNPVDFVGDK | 6 | 2.69 | 1 | 0.094 | 1344 | 1365 | 1368 | 1404 | 5′ | 9 | 12 |
| 14 | cg | k | r2 | LLASLPQPTVVPSTMPTISVRSGVLAGCLIGWWKPK | 2 | 2.00 | 1 | 0.592 | 11103 | 11058 | 11163 | 11106 | 3′ | 16 | 20 |
| 15 | cg | t | r0 | TDNTNHHLTGSAIMTMTAPVK | 540 | 4.89 | 0 | 0.653 | 11625 | 11601 | 11661 | 11628 | 3′ | 11 | 10 |
| 16 | cg | no | r1 | TSQTDLLTDPPITYEFLWAFSVNK$ | 8 | 2.03 | 0 | 0.592 | 12150 | 12126 | 12195 | 12153 | 3′ | 9 | 14 |
| 17 | ct | m | f1 | MWEDLMVEAMNSLSSATVGR | 880 | 4.49 | 0 | 0.837 | 1995 | 2019 | 2022 | 2052 | 5′ | 9 | 11 |
| 18 | ct | e | r0 | GMGPMAYLASLALKENMVNNAEGFK | 642 | 2.47 | 1 | 0.182 | 4233 | 4209 | 4206 | 4161 | 5′ | 9 | 16 |
| 19 | ct | t | f0 | NPSLSISVPSTRHVSMPITISSIPPQTTEMCLMK | 3 | 2.54 | 1 | 0.866 | 4290 | 4350 | 4251 | 4287 | 3′ | 21 | 13 |
| 20 | ct | a | r2 | GVNWAKMNIAGYESSYNEQR | 5 | 2.76 | 1 | 0.352 | 10512 | 10485 | 10542 | 10515 | 3′ | 10 | 10 |
| 21 | ct | m | r0 | AMMGDCAVCGTEMMSMCIK | 13 | 2.03 | 0 | 0.42 | 13914 | 13890 | 13887 | 13860 | 5′ | 10 | 9 |
| 22 | ct | v | r0 | NVVWSVAVAAMMKGGVGVGMGGHMEMK | 31 | 3.31 | 1 | 0.393 | 15321 | 15279 | 15276 | 15240 | 5′ | 14 | 13 |
| 23 | ct | y | f1 | DVSGPSSPSSSLMTLTLFSPDLLGDPDNYTLANPLNTPPY | 2 | 2.27 | 0 | 0.724 | 15714 | 15801 | 15684 | 15711 | 3′ | 30 | 10 |
| 24 | gt | q | r2 | NNLFSLYCYLFQLWMMDPEHMNSMALK | 1 | 2.55 | 0 | 0.464 | 13329 | 13287 | 13365 | 13332 | 3′ | 16 | 11 |
| 25 | ac gt | s | f0 | MVGSFMGSGDKPTEPGDSFGEPRSEAGPGPGSTLQSAR | 38 | 2.43 | 1 | 0.459 | 1995 | 2049 | 2052 | 2109 | 5′ | 19 | 19 |
| 26 | ac gt | h | r2 | WSSSLAAPSAFVLVGMSSRHSLLVCGTHVYFFGHNWNK | 1 | 2.71 | 1 | 0.218 | 4428 | 4389 | 4500 | 4431 | 3′ | 14 | 24 |
| 27 | ac gt | s | f2 | SGWVEWSRHSVLLLLSLPVLAAGITMLLTDR | 8 | 3.85 | 1 | 1.000 | 6591 | 6648 | 6558 | 6588 | 3′ | 21 | 10 |
| 28 | ac gt | y | f0 | EATASSAGNDASYDGQSGKDSQATPYTKPTPK | 9 | 2.77 | 1 | 0.425 | 7221 | 7254 | 7161 | 7218 | 3′ | 12 | 20 |
| 29 | ac gt | x | r1 | GDKLFYDXGLLWGAQAGMVR | 435 | 4.06 | 1 | 0.901 | 7503 | 7479 | 7476 | 7446 | 5′ | 9 | 11 |
| 30 | ac gt | x | r1 | RPLSPXGASLWSSVLXTYLR | 129 | 5.31 | 0 | 0.516 | 7476 | 7452 | 7509 | 7479 | 3′ | 9 | 11 |
| 31 | ac gt | n | f0 | VMVTDLLQKSWSPHSYNNYITNR | 6 | 2.39 | 1 | 0.971 | 8157 | 8193 | 8127 | 8154 | 3′ | 13 | 10 |
| 32 | ac gt | n | f2 | SNALNNAGKNAEGHYSSSPNNK | 1 | 2.52 | 1 | 1.000 | 8559 | 8583 | 8520 | 8556 | 3′ | 9 | 13 |
| 33 | ac gt | c | f1 | TPGVVPEPAPAANVHSSCPPCPWLPCFPPSLPPSLTLTK | 15 | 2.75 | 0 | 0.296 | 12564 | 12624 | 12510 | 12561 | 3′ | 22 | 17 |
| 34 | ac gt | no | f0 | SLKQNWDFSFNSSTMVVAGIFLLIR | 1 | 2.11 | 1 | 1.000 | 13299 | 13338 | 13266 | 13296 | 3′ | 14 | 11 |
| 35 | ac gt | x | f1 | EMHLCSXEDSRAHNTWGXLK | 13 | 2.02 | 1 | 0.989 | 16728 | 16752 | 16698 | 16725 | 3′ | 10 | 10 |
| 36 | ag ct | k | r2 | SLAPSGWSLLNLTNPLFSSMNLPTILLHKR | 16 | 4.31 | 1 | 0.208 | 1728 | 1704 | 1788 | 1731 | 3′ | 11 | 19 |
| 37 | ag ct | d | r1 | LGDDWLEDMGNSNQNQLK | 3 | 2.08 | 0 | 0.899 | 3426 | 3399 | 3396 | 3375 | 5′ | 9 | 9 |
| 38 | ag ct | no | f2 | WALFLSGTDSSSVSLAPLAATGSWGGLNQTQLR | 7 | 2.96 | 0 | 0.143 | 5043 | 5076 | 4980 | 5040 | 3′ | 12 | 21 |
| 39 | ag ct | y | f0 | NPPYTWSDYMSIFCFVVCLGGLR | 15 | 2.08 | 0 | 0.875 | 7485 | 7515 | 7518 | 7554 | 5′ | 10 | 13 |
| 40 | ag ct | e | r1 | YVGVEDESAVTNTSTNLTLPTIGQPSNGKK | 2 | 2.14 | 1 | 0.472 | 7809 | 7779 | 7776 | 7719 | 5′ | 10 | 20 |
| 41 | ag ct | q | r2 | GDACWGPVPSQLGGQGQAGVVKGLQGLHQQGGPQNGGR | 1 | 2.24 | 1 | 0.804 | 9456 | 9387 | 9384 | 9342 | 5′ | 23 | 15 |
| 42 | ag ct | no | f1 | AHVEAPIAGSMVLAVTSPGSNNR | 37 | 3.74 | 0 | 1.000 | 11604 | 11643 | 11646 | 11670 | 5′ | 14 | 9 |
| 43 | ag ct | v | r1 | RSPLPGDQVDYVVVHGGMSVQFLWAFSVNK | 34 | 3.08 | 1 | 1.000 | 12180 | 12126 | 12213 | 12183 | 3′ | 19 | 11 |
| 44 | ag ct | f | f1 | FNPFFGFVGPITKPTLNFNK | 914 | 3.51 | 0 | 0.423 | 14874 | 14904 | 14847 | 14871 | 3′ | 11 | 9 |
| 45 | ag ct | v | r2 | HVHPEPSDEVAAYGANSIRCVGVGVVVMLVR | 1 | 3.23 | 1 | 0.721 | 15015 | 14979 | 15069 | 15018 | 3′ | 13 | 18 |
| 46 | at cg | no | f2 | SSLRPYTKCVVFLASEEVK | 4 | 2.41 | 1 | 1.000 | 3135 | 3129 | 3126 | 3096 | 5′ | 9 | 10 |
| 47 | at cg | e | r2 | QAEVFLSLQSSSQNHCFMQHISSGESASYVVPEK | 215 | 3.23 | 0 | 0.224 | 3858 | 3822 | 3921 | 3861 | 3′ | 13 | 21 |
| 48 | at cg | a | r0 | FEDNKWDSFIDFYQTYFLGLAGNAGDCNGYGDMSYK | 1 | 2.82 | 1 | 1.000 | 4074 | 4002 | 4107 | 4077 | 3′ | 24 | 12 |
| 49 | at cg | e | f2 | GISWPKLDEEGGGPFEAGEAPAGLK | 59 | 3.82 | 1 | 0.169 | 5847 | 5874 | 5799 | 5844 | 3′ | 9 | 16 |
| 50 | at cg | k | f1 | SIAGVDVAMAVSGTKTLYLLHSNTHHNR | 1 | 3.31 | 1 | 0.886 | 6201 | 6231 | 6147 | 6198 | 3′ | 10 | 18 |
| 51 | at cg | r | f2 | WLPWLGCSCGWCRLITSTPTYFPHYSR | 1 | 2.49 | 1 | 0.941 | 8070 | 8097 | 8022 | 8067 | 3′ | 11 | 16 |
| 52 | at cg | c | r2 | CYLVGAFHCNLHNQENCK | 27 | 3.92 | 0 | 0.690 | 8247 | 8223 | 8220 | 8196 | 5′ | 9 | 9 |
| 53 | at cg | e | r1 | AGEGLLEVWKASEPNSAVAK | 8 | 3.17 | 1 | 0.171 | 9072 | 9042 | 9039 | 9012 | 5′ | 10 | 10 |
| 54 | at cg | e | f1 | EIFLSLLPQVGSGMGGESSR | 11 | 3.57 | 0 | 0.249 | 9648 | 9669 | 9672 | 9705 | 5′ | 9 | 11 |
| 55 | at cg | h | f2 | GFLCIKLSCVGGCPHLLASSLYYFLTK$ | 8 | 2.08 | 1 | 0.935 | 11037 | 11070 | 10992 | 11034 | 3′ | 13 | 17 |
| 56 | at cg | x | r0 | DGGNXGSQGXGAMSSHVPMMKMNLNVVLK$ | 38 | 2.25 | 1 | 0.766 | 12321 | 12291 | 12288 | 12237 | 5′ | 12 | 17 |
| 57 | at cg | r | f0 | RPRLTSLPSLLNDINTILWSGGSAGSVNMGSVGEFVGR | 1 | 2.94 | 1 | 0.124 | 15687 | 15735 | 15738 | 15798 | 5′ | 18 | 20 |
| 58 | acg | n | r2 | NTTTLSRTLNVGAVMNNVMVDVAGFNGSLVK | 60 | 2.68 | 1 | 1.000 | 3315 | 3255 | 3345 | 3318 | 3′ | 21 | 10 |
| 59 | acg | e | f0 | SEHTPQLPTETTSSALSDRR | 159 | 3.52 | 1 | 0.141 | 5085 | 5109 | 5112 | 5142 | 5′ | 10 | 10 |
| 60 | acg | g | r0 | WGGSTTNGGEPTGGSTLVGGEYKLQGDR | 173 | 2.52 | 1 | 0.351 | 6483 | 6450 | 6531 | 6486 | 3′ | 12 | 16 |
| 61 | acg | q | r2 | QLVEQPKDTVEWQDMVEVGYNVVR | 3 | 5.28 | 1 | 0.401 | 6891 | 6852 | 6924 | 6894 | 3′ | 13 | 11 |
| 62 | acg | t | f2 | TSKPHPTTTPPPSSSTPLQGLQCGGVRGVQAHQGAGHVQGR | 73 | 3.03 | 1 | 0.876 | 13167 | 13215 | 13218 | 13287 | 5′ | 17 | 24 |
| 63 | acg | t | f2 | TSKPHPTTTPPPSSSTQISPITCGGVRGVQAHQGAGHVQGR | 29 | 2.99 | 1 | 0.866 | 13167 | 13230 | 13233 | 13290 | 5′ | 22 | 19 |
| 64 | acg | no | f2 | GKQEPGLEQLCASSAALGEIPLPNNNPPLPK | 17 | 2.36 | 1 | 0.236 | 13989 | 14022 | 13932 | 13986 | 3′ | 12 | 19 |
| 65 | acg | e | f0 | GLPQHQVHHKPHKPHYETHTQQK | 2 | 2.05 | 0 | 1.000 | 14862 | 14901 | 14835 | 14859 | 3′ | 14 | 9 |
| 66 | acg | e | f0 | GLPQHQVHHQPHKPHYETHTQQK | 2 | 2.05 | 0 | 1.000 | 14865 | 14901 | 14835 | 14862 | 3′ | 13 | 10 |
| 67 | acg | a | f1 | GGFSGPSQILIATILCSFMGK | 4 | 2.24 | 0 | 0.319 | 16266 | 16305 | 16242 | 16263 | 3′ | 12 | 9 |
| 68 | act | e | r2 | GENAGELEGWTATLSCSSPEGPTTLGPLR | 6 | 2.59 | 0 | 0.156 | 3228 | 3186 | 3273 | 3231 | 3′ | 14 | 15 |
| 69 | act | t | r1 | KPAGASPAFFPGGGTSTLKPVDTGATLLTMEGETVSPGSVVK$ | 2 | 2.04 | 0 | 1.000 | 5598 | 5511 | 5508 | 5472 | 5′ | 30 | 12 |
| 70 | act | k | f2 | NLPILLLTNVEPQPFPPTPK | 27 | 4.28 | 0 | 0.948 | 11097 | 11118 | 11064 | 11094 | 3′ | 9 | 11 |
| 71 | act | k | f2 | NLPILLLTNVDPQPFPPTPK | 197 | 4.97 | 0 | 1.000 | 11097 | 11118 | 11064 | 11094 | 3′ | 9 | 11 |
| 72 | act | n | f2 | WPLQCRQMSTTSSNFCHFNPNNNPPLPK | 3 | 2.93 | 1 | 0.970 | 13998 | 14022 | 13941 | 13995 | 3′ | 9 | 19 |
| 73 | agc | r | f2 | DQPNPLRPCPAPLTDAMAIR | 45 | 2.19 | 0 | 0.272 | 3945 | 3969 | 3972 | 4002 | 5′ | 10 | 10 |
| 74 | agc | k | r1 | NNSSPIPVPVSSMVMPAAKTGK | 365 | 3.86 | 1 | 0.382 | 6369 | 6336 | 6396 | 6372 | 3′ | 13 | 9 |
| 75 | agc | v | r2 | FQLQSMPVLGSVGGVGGAMGVMR | 125 | 4.64 | 0 | 0.958 | 8157 | 8115 | 8184 | 8160 | 3′ | 14 | 9 |
| 76 | agc | no | r2 | TSYLLLQQPLLWWLCWFKLCVFWK | 5 | 2.01 | 1 | 0.163 | 10749 | 10716 | 10785 | 10752 | 3′ | 12 | 12 |
| 77 | agc | q | r1 | MQQAAFALQWMLLWGQDWQQQWMR | 8 | 2.16 | 0 | 0.264 | 12432 | 12390 | 12459 | 12435 | 3′ | 15 | 9 |
| 78 | agc | w | r2 | MSSTCGNDISMSRISGLFSAWGWK | 4 | 2.04 | 1 | 0.739 | 13098 | 13068 | 13137 | 13101 | 3′ | 11 | 13 |
| 79 | agc | q | f0 | LQQMMQQADVLVAGIFLLIR | 13 | 3.77 | 0 | 0.353 | 13314 | 13338 | 13281 | 13311 | 3′ | 9 | 11 |
| 80 | agc | v | f0 | IVAFSTSSQLGLMVLEVPVGVK | 260 | 4.16 | 0 | 0.296 | 13458 | 13488 | 13491 | 13515 | 5′ | 13 | 9 |
| 81 | agc | d | r1 | FLVDLGLGGVGAGFGSDEGLDDGLCGVWCDFMTVSLLMNK | 2 | 2.80 | 0 | 0.029 | 13971 | 13887 | 13884 | 13854 | 5′ | 30 | 10 |
| 82 | agt | a | f0 | TDAQSGGASAYKAHENILLR | 1 | 2.25 | 1 | 1.000 | 2343 | 2370 | 2313 | 2340 | 3′ | 10 | 10 |
| 83 | agt | n | f2 | LIYSTSITLLPMTGGNGEGMR | 194 | 4.00 | 0 | 0.586 | 5442 | 5475 | 5478 | 5502 | 5′ | 12 | 9 |
| 84 | agt | d | f2 | LIYSTSITLLPMTGGDGEGMR | 61 | 3.96 | 0 | 0.460 | 5442 | 5475 | 5478 | 5502 | 5′ | 11 | 10 |
| 85 | agt | r | r2 | NGVSSSGGVEEGGVEVAVCLLLCVEWWLVRVCLVLLVR | 1 | 2.41 | 1 | 0.180 | 6423 | 6369 | 6366 | 6312 | 5′ | 20 | 18 |
| 86 | agt | g | r2 | AASCGPPSCLPEVGINGGGNGMISTAAGGPGIVGAMNEANG* | 1 | 2.29 | 0 | 0.275 | 8493 | 8409 | 8526 | 8496 | 3′ | 30 | 11 |
| 87 | agt | a | r2 | AAEGNSYAEEFYGEADAGGGYAVEAATAWGGPPLAEAVPR | 1 | 2.43 | 0 | 0.518 | 10143 | 10068 | 10065 | 10026 | 5′ | 25 | 15 |
| 88 | agt | d | r0 | NYSSAMGACQGGSDESNDDGSGVCVWFARGPVVAAPGAVDR | 6 | 2.55 | 1 | 0.953 | 13566 | 13491 | 13488 | 13446 | 5′ | 26 | 15 |
| 89 | agt | y | f2 | RIRPVEVVGAIPHYFLLQYPHHR* | 1 | 2.81 | 1 | 0.173 | 13713 | 13749 | 13680 | 13710 | 3′ | 12 | 11 |
| 90 | atc | h | f1 | HPLWNLHVEGGFSSNTCAVRPAVMGNVESYMHKHK | 12 | 2.55 | 1 | 0.124 | 1413 | 1452 | 1455 | 1515 | 5′ | 15 | 20 |
| 91 | atc | q | f1 | GTLTVQQQHNMPPTFLGNAR | 61 | 3.19 | 0 | 0.659 | 2616 | 2643 | 2646 | 2673 | 5′ | 10 | 10 |
| 92 | atc | v | r2 | GLVIVVVKALGSVGGVGGAMGVMR | 429 | 4.41 | 1 | 0.299 | 8157 | 8115 | 8187 | 8160 | 3′ | 14 | 10 |
| 93 | atc | v | r2 | GLVIVVVKAVGSVGGVGGAMGVMR | 1057 | 4.48 | 1 | 0.140 | 8160 | 8115 | 8187 | 8163 | 3′ | 14 | 10 |
| 94 | atc | x | r1 | MXLMLMIALVAXEXIQMVVLFSVMAGMLGVVGWCR$ | 1 | 2.16 | 0 | 0.818 | 13263 | 13233 | 13335 | 13266 | 3′ | 11 | 24 |
| 95 | atc | k | r0 | FNYAFLGWGDWLLLWNYHMGMKVK | 17 | 3.81 | 1 | 1.000 | 14049 | 14022 | 14019 | 13980 | 5′ | 10 | 14 |
| 96 | atc | q | r2 | LWLCSKGGQWLQLGFVMNQFLMNDPK | 35 | 2.60 | 1 | 0.630 | 15408 | 15381 | 15378 | 15333 | 5′ | 11 | 15 |
| 97 | atg | n | f0 | TLGQGVAHEVANNGLHLRPLFSSCTR | 1 | 2.13 | 0 | 0.076 | 1344 | 1389 | 1392 | 1419 | 5′ | 17 | 9 |
| 98 | atg | d | r1 | EPMMHQVSMGKKPVSGGDPPDEDDVDGIK | 14 | 2.57 | 1 | 0.129 | 5061 | 5007 | 5088 | 5064 | 3′ | 19 | 10 |
| 99 | atg | no | f0 | TMASSSPPSVPPAPAGSASASVTVASPPLALVAPVR$ | 13 | 2.42 | 0 | 1.000 | 5376 | 5409 | 5412 | 5481 | 5′ | 9 | 27 |
| 100 | atg | no | f2 | GASFLFIWNSLYLLFGAWAGVLGTALSLLIRAELGQPGNL | 1 | 2.24 | 1 | 1.000 | 6054 | 6141 | 6024 | 6051 | 3′ | 30 | 10 |
| 101 | atg | m | r0 | IMRMGAFGIGNMSGENTSTK | 65 | 2.86 | 1 | 1.000 | 6291 | 6264 | 6261 | 6234 | 5′ | 10 | 10 |
| 102 | atg | p | f0 | APPTALVGTDSPHSTEAMWNDLLQCSEPPDSSFFSPPVA | 10 | 2.64 | 0 | 0.564 | 6987 | 7074 | 6960 | 6984 | 3′ | 30 | 9 |
| 103 | atg | k | r1 | KPYTLPMESMNPFCSHSKAK | 51 | 2.17 | 1 | 1.000 | 10284 | 10263 | 10314 | 10287 | 3′ | 10 | 10 |
| 104 | atg | n | r1 | GGAYQGNQSNNLLGGGLVVGWGLDNRLEGLFVVGLMSWSVG | 3 | 3.02 | 1 | 0.924 | 12936 | 12846 | 12969 | 12939 | 3′ | 30 | 11 |
| 105 | atg | x | r1 | EWAEVSSCGEEGXGGADAASEEPTKTTGER | 11 | 2.51 | 1 | 0.991 | 14826 | 14784 | 14781 | 14739 | 5′ | 10 | 19 |
| 106 | cgt | q | r1 | GLYWWDQQYGSGQGGWSLASPLDLGAQWTQGVGFR | 1 | 2.15 | 0 | 1.000 | 228 | 189 | 186 | 126 | 5′ | 14 | 21 |
| 107 | cgt | x | f0 | HPKPKPWEMXLXIPLXIXLLVQXGTALWTLGK | 1 | 2.72 | 0 | 0.632 | 2103 | 2166 | 2073 | 2100 | 3′ | 22 | 10 |
| 108 | cgt | m | r0 | LLQSDHTAFGSAPMSPQPK | 2 | 2.13 | 0 | 0.217 | 2604 | 2577 | 2631 | 2607 | 3′ | 10 | 9 |
| 109 | cgt | a | f2 | AGASEGMTSYMMCLHTHYNLQHSPSNLANMSDK | 6 | 2.06 | 0 | 0.034 | 4278 | 4347 | 4251 | 4275 | 3′ | 23 | 10 |
| 110 | cgt | t | r2 | RPPSGWPSCTSTLVSGATTTTGRGAGVTVETTECGSSTDNM | 6 | 2.51 | 1 | 0.584 | 4989 | 4902 | 5028 | 4992 | 3′ | 30 | 11 |
| 111 | cgt | no | r0 | LFLGMCLKQENPVMMSGLK | 393 | 4.09 | 1 | 0.162 | 12066 | 12033 | 12093 | 12069 | 3′ | 10 | 9 |
| 112 | cgt | t | f0 | VFLLTMTFNQNITLWIWQHIASTGHPGMNATMSCK | 2 | 2.14 | 0 | 0.703 | 12330 | 12384 | 12387 | 12435 | 5′ | 19 | 17 |
| 113 | cgt | f | f2 | MNGTEGHVVHVPDGVASMIYPTLQLMFPTTNSPSK | 8 | 2.57 | 0 | 0.942 | 13107 | 13161 | 13059 | 13104 | 3′ | 19 | 16 |
| 114 | ctg | v | r0 | VFVSLSLVSPFWLNPASTVAVNVNGYNEEVEVGHGYVVK | 1 | 2.98 | 0 | 1.000 | 3126 | 3078 | 3192 | 3129 | 3′ | 17 | 22 |
| 115 | ctg | e | f1 | QSHMKSPEPVGDEEEDEER | 26 | 4.14 | 1 | 0.032 | 3783 | 3807 | 3810 | 3837 | 5′ | 9 | 10 |
| 116 | ctg | k | r2 | DQVRPLVLCMVMLYFTIHLLHAYK | 8 | 2.55 | 0 | 0.480 | 8601 | 8571 | 8568 | 8532 | 5′ | 12 | 12 |
| 117 | ctg | k | f0 | MNNKVFLVFVQTTIPLYLK | 791 | 3.61 | 1 | 0.130 | 14001 | 14025 | 13971 | 13998 | 3′ | 9 | 10 |
| 118 | ctg | no | f2 | SPSSMYPNNKLEEGLYELK | 70 | 4.56 | 1 | 0.048 | 15915 | 15945 | 15948 | 15972 | 5′ | 10 | 9 |
| 119 | acgt | t | f1 | GGAGGTPAATGTRTPSAGSDSVSTDFTQPTTSTTTPTNLK* | 5 | 2.24 | 1 | 0.825 | 2520 | 2466 | 2586 | 2523 | 3′ | 18 | 22 |
| 120 | acgt | a | r1 | FVKAALFLLAGTYPSLGAR | 79 | 4.05 | 1 | 0.342 | 2937 | 2913 | 2910 | 2883 | 5′ | 10 | 9 |
| 121 | acgt | d | r1 | MVEDMTGWADGLISTGDVDPTFSGVPKLSGGSAK | 6 | 2.88 | 1 | 0.287 | 4305 | 4236 | 4233 | 4206 | 5′ | 23 | 11 |
| 122 | acgt | f | r2 | EEMLDGSFCGTFVFGGPVLALFSVMR | 1 | 2.20 | 0 | 0.163 | 4452 | 4416 | 4413 | 4371 | 5′ | 14 | 12 |
| 123 | acgt | no | r2 | VPRQALVPFEVNEASYDGK | 141 | 3.13 | 1 | 0.592 | 6930 | 6906 | 6960 | 6933 | 3′ | 9 | 10 |
| 124 | acgt | no | r2 | VPRQALVPFDVNEASYDGK | 388 | 3.16 | 1 | 0.496 | 6930 | 6906 | 6960 | 6933 | 3′ | 9 | 10 |
| 125 | acgt | no | f1 | SNFLPTTLSRPIRNAPTLLGLGVMHAAHPAGQQMLEQAK | 29 | 2.97 | 1 | 1.000 | 7287 | 7350 | 7353 | 7404 | 5′ | 21 | 18 |
| 126 | acgt | q | r1 | HGQAMLALPVLDPSVVVLGGCQGVGGK | 1 | 2.74 | 0 | 0.031 | 10857 | 10833 | 10911 | 10860 | 3′ | 9 | 18 |
| 127 | acgt | no | r2 | VVGGVGWVPLAWLSLDMLQR | 167 | 3.64 | 0 | 0.555 | 14448 | 14424 | 14481 | 14451 | 3′ | 9 | 11 |
| 128 | acgt | a | r2 | WMSGALILLGGAFCVLGSFRGGVGFVLLPGDVMADAGVER* | 4 | 2.37 | 1 | 1.000 | 14772 | 14718 | 14715 | 14652 | 5′ | 18 | 22 |
| 129 | actg | d | r1 | SSYKDPFAEDADLDNDIALLSLGDLVPLVK | 149 | 3.81 | 1 | 0.469 | 2733 | 2682 | 2769 | 2736 | 3′ | 17 | 13 |
| 130 | actg | q | r2 | GMGQGVHSQQAMVQAKVGAVMQQVMVDVAGGQNVGQPQGR | 2 | 2.36 | 1 | 0.409 | 3363 | 3276 | 3273 | 3246 | 5′ | 30 | 10 |
| 131 | actg | s | f0 | NSVCSDGSARAVSPLAPGLSHK | 25 | 2.75 | 1 | 0.560 | 3273 | 3303 | 3306 | 3315 | 5′ | 10 | 12 |
| 132 | actg | s | f0 | NSVCSDGSARSSSPLAPGLSHK | 23 | 3.21 | 1 | 1.000 | 3273 | 3309 | 3312 | 3339 | 5′ | 12 | 10 |
| 133 | actg | s | r0 | MIMSAWSWKVMSSSMMETSMVEHLLDMIEIRPR | 6 | 2.41 | 1 | 0.948 | 7527 | 7482 | 7479 | 7431 | 5′ | 17 | 16 |
| 134 | actg | no | f2 | SLSPFMITPSSVGVGVMVAVER | 59 | 3.28 | 0 | 0.884 | 7767 | 7794 | 7797 | 7830 | 5′ | 11 | 11 |
| 135 | actg | t | r0 | TPKEQEIGEATVGTGIFNLTAK | 2 | 2.42 | 1 | 1.000 | 8778 | 8748 | 8811 | 8781 | 3′ | 11 | 11 |
| 136 | actg | no | f2 | NQMIQALLITILLGLYFTLLSIVTAGTVFGLR | 1 | 2.29 | 0 | 0.690 | 9834 | 9894 | 9897 | 9930 | 5′ | 20 | 12 |
| 137 | actg | f | r1 | SNAFGESKFTPTETMADTMAAFFFEYCGK | 1 | 2.42 | 1 | 0.403 | 11400 | 11367 | 11451 | 11403 | 3′ | 12 | 17 |
| 138 | actg | r | r2 | WGSSNHEHGGAGCLMGMVQGR | 571 | 4.35 | 0 | 0.165 | 11481 | 11460 | 11517 | 11484 | 3′ | 9 | 12 |
| 139 | actg | k | r2 | WGSSNHEHGGAGGLMGMVQGKGK | 67 | 4.67 | 1 | 0.973 | 11478 | 11454 | 11517 | 11481 | 3′ | 10 | 13 |
| 140 | actg | d | r0 | AMLLDMGAWVSKVETWVDAR | 9 | 4.42 | 1 | 1.000 | 12186 | 12153 | 12150 | 12126 | 5′ | 11 | 9 |
| 141 | actg | d | r0 | AMLLDMGAWVSQVETWVDAR | 9 | 4.42 | 0 | 1.000 | 12186 | 12153 | 12150 | 12123 | 5′ | 10 | 10 |
| 142 | actg | no | f2 | VSAASSGFYRPLPNNNPPLPK | 81 | 3.67 | 0 | 1.000 | 13992 | 14022 | 13962 | 13989 | 3′ | 11 | 10 |
| 143 | agct | k | r2 | SKTLLLMWTLILIQSTSGK | 110 | 3.76 | 1 | 0.377 | 2757 | 2730 | 2727 | 2703 | 5′ | 10 | 9 |
| 144 | agct | s | f0 | ASLSVALDPSGSTNTSLTAKHPNQLASIYFSR | 2 | 2.24 | 1 | 0.987 | 5775 | 5832 | 5730 | 5772 | 3′ | 20 | 12 |
| 145 | agct | no | f0 | KAPNPCLAICALDACMLEK | 1222 | 3.24 | 1 | 0.448 | 5931 | 5955 | 5958 | 5985 | 5′ | 9 | 10 |
| 146 | agct | s | f0 | GGPAGSVFWATIQANPMASMTFSKSYSK | 4 | 2.67 | 1 | 0.429 | 7608 | 7653 | 7572 | 7605 | 3′ | 16 | 12 |
| 147 | agct | h | r0 | TLLNKTSPTFTYLSGEAHLTCHGEK* | 4 | 2.19 | 1 | 0.342 | 13599 | 13569 | 13641 | 13602 | 3′ | 11 | 14 |
| 148 | agtc | h | f0 | SLLPSLSTQHHRYSGWVPGGSGLDIHAPK | 1 | 2.81 | 1 | 0.420 | 2445 | 2472 | 2475 | 2529 | 5′ | 12 | 17 |
| 149 | agtc | r | f2 | AATDDERPTTTGLNSSTTTLLLSR | 5 | 2.66 | 0 | 0.303 | 5214 | 5247 | 5163 | 5211 | 3′ | 11 | 13 |
| 150 | agtc | e | f1 | LTEPLTNGESSWEKASGSMVLAAVLLK$ | 155 | 2.13 | 1 | 0.932 | 11628 | 11658 | 11580 | 11625 | 3′ | 11 | 16 |
| 151 | agtc | m | f0 | IVAFSTSSQLGLMASEALAGMK | 168 | 3.53 | 0 | 0.790 | 13458 | 13494 | 13497 | 13521 | 5′ | 13 | 9 |
| 152 | agtc | w | f0 | IVAFSTSSQLGLMASEALAGWK | 434 | 4.72 | 0 | 0.715 | 13458 | 13494 | 13497 | 13521 | 5′ | 13 | 9 |
| 153 | agtc | d | f0 | IVAFSTSSQLGLMASEALAGDK | 448 | 4.33 | 0 | 0.817 | 13458 | 13494 | 13497 | 13521 | 5′ | 12 | 10 |
| 154 | agtc | r | r1 | SLLPFVLFTFTPWSTLGLSMFLWVLGRGGLLFGR* | 1 | 3.40 | 1 | 1.000 | 13758 | 13725 | 13821 | 13761 | 3′ | 13 | 21 |
| 155 | agtc | no | r2 | VAPVMSLIWFVLLPVSGPILEWCGRLMK | 16 | 3.06 | 1 | 0.437 | 14934 | 14907 | 14991 | 14937 | 3′ | 9 | 19 |
| 156 | atcg | k | r2 | ATKTVGGVFGQTNQSPDPK* | 1 | 2.68 | 1 | 0.251 | 321 | 294 | 291 | 267 | 5′ | 10 | 9 |
| 157 | atcg | y | f1 | SVGGGSAGYCVCGAAWGLGEPTKPQYHPPQFMYLTSSK | 8 | 2.37 | 0 | 0.517 | 555 | 609 | 498 | 552 | 3′ | 19 | 19 |
| 158 | atcg | e | r0 | VISSEFIMQSQSPKHELEK | 42 | 3.08 | 1 | 0.103 | 1629 | 1605 | 1602 | 1575 | 5′ | 10 | 9 |
| 159 | atcg | a | f2 | LYSQAFNSSSAQHTHGVGGCGVHWVRFEFK | 1 | 2.25 | 1 | 0.669 | 3324 | 3369 | 3372 | 3411 | 5′ | 15 | 25 |
| 160 | atcg | no | f2 | LMPPLCKIHHESVALLVR | 20 | 2.55 | 1 | 1 | 3936 | 3912 | 3909 | 3885 | 5′ | 9 | 9 |
| 161 | atcg | g | r1 | EKNQAVPEGPSMFISGPTQVK | 3 | 2.47 | 1 | 0.573 | 4383 | 4353 | 4413 | 4386 | 3′ | 11 | 10 |
| 162 | atcg | g | r1 | EKNQAVPEGLAMFISGPTQVK | 15 | 2.81 | 1 | 0.57 | 4380 | 4353 | 4413 | 4383 | 3′ | 11 | 10 |
| 163 | atcg | g | r1 | EKNQAVPEGLSMFISGPTQVK | 20 | 2.80 | 1 | 0.57 | 4383 | 4353 | 4413 | 4386 | 3′ | 11 | 10 |
| 164 | atcg | no | f0 | AHTPKMLVMGPGLLPSGQGLGR | 27 | 2.13 | 1 | 0.472 | 4503 | 4527 | 4530 | 4566 | 5′ | 10 | 12 |
| 165 | atcg | x | r1 | SEASASGSAKAAHDHLDDHPMMXLLFFVNSSMMAHLGK | 1 | 2.79 | 1 | 0.34 | 5115 | 5064 | 5175 | 5118 | 3′ | 18 | 20 |
| 166 | atcg | q | r2 | QDCCDQDGSDEDPSNQNPQPAPKQER | 1 | 2.42 | 1 | 0.309 | 6315 | 6282 | 6279 | 6240 | 5′ | 12 | 14 |
| 167 | atcg | v | r2 | GPVTVQAKVVGSVGGVGGAMGVMR | 203 | 4.83 | 1 | 0.074 | 8160 | 8115 | 8187 | 8163 | 3′ | 15 | 9 |
| 168 | atcg | v | r2 | GPVTVQAKVLGSVGGVGGAMGVMR | 181 | 4.59 | 1 | 0.082 | 8157 | 8115 | 8187 | 8160 | 3′ | 14 | 10 |
| 169 | atcg | a | f2 | AASHPVPVPMTLLMLGLLTNTLTMYQWWR | 24 | 2.66 | 0 | 0.053 | 9477 | 9537 | 9450 | 9474 | 3′ | 19 | 10 |
| 170 | atcg | a | r0 | AMTLHAHAGAMFSEPAVLWVAISAMSAGAEPTAVANAK | 2 | 2.44 | 0 | 0.693 | 11196 | 11115 | 11226 | 11199 | 3′ | 28 | 10 |
| 171 | atcg | q | r2 | GDAGEMLLVNAGLLGAQFLLASK | 28 | 3.47 | 0 | 0.068 | 13719 | 13695 | 13692 | 13653 | 5′ | 10 | 13 |
| 172 | atcg | e | r2 | GDAGEMLLVIAGLLGAEFLLASK | 29 | 3.54 | 0 | 0.147 | 13719 | 13695 | 13692 | 13653 | 5′ | 10 | 13 |
| 173 | atcg | t | r0 | SAEHSLGAGYHSGLMWGGVFKGLATVTLSGSPTTSGENT | 1 | 2.93 | 1 | 0.885 | 15546 | 15456 | 15573 | 15549 | 3′ | 30 | 9 |
| 174 | atgc | c | f2 | NIPFLLFGVNSCCVIPSCNMPSACWINCKCLCK | 1 | 2.36 | 1 | 0.239 | 1872 | 1911 | 1815 | 1869 | 3′ | 13 | 19 |
| 175 | atgc | e | f0 | SVESMLLGEENNFAEEAKAK | 641 | 3.52 | 1 | 0.148 | 1902 | 1929 | 1872 | 1899 | 3′ | 11 | 9 |
| 176 | atgc | w | f0 | WDISQGKTFAVILNLVLYPHPPK | 23 | 3.62 | 1 | 0.851 | 3240 | 3270 | 3204 | 3237 | 3′ | 11 | 12 |
| 177 | atgc | k | f1 | HYLYDMSPLNGIENHGKK | 154 | 3.19 | 1 | 0.419 | 4272 | 4293 | 4296 | 4323 | 5′ | 9 | 9 |
| 178 | atgc | h | f1 | LMHHHYKSSAHHVHSPMIVHHNNYQYK* | 35 | 2.40 | 1 | 0.591 | 6501 | 6522 | 6444 | 6498 | 3′ | 9 | 18 |
| 179 | atgc | e | r1 | IINITAVEENPSGRSSLHK | 32 | 3.62 | 1 | 0.040 | 7155 | 7131 | 7128 | 7101 | 5′ | 10 | 9 |
| 180 | atgc | e | r1 | IINITAVEEIPSGRSSLHK | 42 | 4.38 | 1 | 0.156 | 7155 | 7131 | 7128 | 7101 | 5′ | 10 | 9 |
| 181 | atgc | no | f2 | LLKECLSLASVPATPPYHTFEEPVYMK | 12 | 2.48 | 1 | 0.681 | 7524 | 7560 | 7482 | 7521 | 3′ | 13 | 14 |
| 182 | atgc | f | f1 | AQWLFAFALFLKMPFPFEVMFHMSMK | 5 | 2.10 | 1 | 1.000 | 9006 | 9051 | 9054 | 9081 | 5′ | 16 | 10 |
| 183 | atgc | n | r1 | NYLYYKSYCVSYSTTNNLSFNITK | 14 | 3.54 | 1 | 0.304 | 11298 | 11268 | 11265 | 11229 | 5′ | 12 | 12 |
| 184 | atgc | k | r0 | SKNKPDTNASSNPVMMSGLK | 232 | 3.49 | 1 | 0.793 | 12054 | 12033 | 12087 | 12057 | 3′ | 9 | 11 |
| 185 | atgc | c | f1 | VIFCQMVEFCVMVQVHSDNCADIIEAPLHKMTSK | 1 | 2.13 | 1 | 0.623 | 13428 | 13452 | 13353 | 13425 | 3′ | 9 | 24 |
| 186 | atgc | y | f1 | VVYNGLQAMPEAYSQDFSLLTTFPPHPPSK | 12 | 2.47 | 0 | 0.484 | 13941 | 14001 | 13914 | 13938 | 3′ | 21 | 9 |
The second testable prediction of the hypothesis is that the same polymerase produces regular and swinger-transformed sequences. Hence occasionally, polymerases switch in the midst of replication/transcription, so that part of the sequence follows regular templating rules, and the other, contiguous part, is swinger transformed according to one among the 23 swinger rules. The fact that there are far more RNAs that are entirely swinger transformed than chimeric RNAs [100] suggests that such switches during polymerization are rare, and usually occur before or at the onset of polymerization.
The 16S rRNA gene in the complete mitogenome of Kamimuria wangi is a swinger transformed A ↔ T + C ↔ G DNA sequence, embedded within an otherwise regular insect mitogenome [99]. The reasons why until now only DNA matching this A ↔ T + C ↔ G swinger transformation has been detected, remain unknown. This A ↔ T + C ↔ G exchange rule is also common among chimeric (part regular, part swinger) RNAs [100].
Peptides encoded by chimeric transcripts are detected for the first time here. An example of three peptides and the corresponding DNA, chimeric RNA is described in Fig. 1. Underlined parts are translated after swinger-transformation of the transcribed RNA sequence. When a detected peptide corresponds only to all or part of the underlined amino acid sequence, this peptide is considered a swinger peptide, as the peptides described in an earlier publication [103]. When the detected peptide encompasses part of the underlined, and part of the contiguous amino acid sequence(s) that are not underlined in Fig. 1, the peptide is considered chimeric, because translated from untransformed RNA (the part that is not underlined in Fig. 1), and from RNA that is swinger translated (underlined part in Fig. 1).
1.4. Previously detected swinger RNA
An anonymous reviewer suggested to add, for reader convenience, explanations on how swinger sequences described in previous publications had been detected in GenBank. These methods are not described in the Materials and methods section, as this would be inadequate and confusing: neither results nor analyses beyond those described in earlier publications on RNA were done in the context of the presently described proteomic analyses. The aim here is (chimeric) peptide detection. Following descriptions are only for the convenience of potential readers.
In a first step, the 23 swinger versions of the human mitogenome were transformed in silico. This means that for swinger transformation A ↔ C (as an example), all As in the human mitogenome are replaced by the ‘replace’ function in the software Word by ‘X’. Then all Cs are replaced by A. The last step is to replace all Xs by C, producing a hypothetical, A ↔ C swinger transformed human mitogenome. Similar procedures produce all 23 possible swinger versions of the mitogenome. Each of these is then analyzed by BLASTn [2]. Two types of analyses have been done. The first analyzes (publications by Seligmann on swinger sequences prior to 2016, from 2012 on) search for alignments between the swinger transformed mitogenome and various sequence databases in GenBank, using standard default megablast parameters. This resulted in detecting long, highly similar sequences, as described in [88], [92], [93], [94], for example. Such searches do not yield alignments with nuclear chromosome sequences, but detect about 100 ESTs (expressed sequence tags). The length of the alignments (> 100 nucleotides) and the similarity with the hypothetical swinger-transformed mitogenome versions (> 90%), as these were previously presented (Table 1 in [88], [92], and Table 2 in [93], [94]), are not compatible with randomly obtained results (as tested by simulations based on randomly shuffled swinger mitogenomes in [96] (therein Section 2.2.3.)).
These long EST swinger sequences were then confirmed by sequences detected within sequence read archives (SRA) of the human transcriptome published by Garzon et al. [30]), GenBank SRA entries SRX768406–SRX768440. For these analyses pertaining to short RNA reads (50 nucleotides, RNA seq, Illumina), BLASTn searched for ‘somewhat similar sequences’, using default search parameters, and detected swinger reads as these are described in [103] (therein supplementary data). These results on short swinger reads converge with those obtained by the first, EST-focused search ([103], therein Figure 2). Using the same search tool and criteria as used for the RNA seq reads, the 23 swinger-transformed mitogenome versions also align with nuclear chromosome sequences (they did not align with human nuclear chromosomes when using megablast as for the EST search).
1.5. Nuclear origins of swinger sequences (numts)
Alignments detected with relaxed criteria (BLASTn, see previous section) between swinger mitogenome sequences and each short reads and nuclear chromosome sequences suggest the possibility that swinger reads (but not swinger ESTs) could originate from the cytosol. To some extent, this is not relevant to the main issue at stake, the very existence of swinger polymerizations, but could be relevant because the very large human nuclear genome could by chance be the origin of these alignments, due to its size.
In addition, mitogenome copies (called numts, [51]) are inserted within nuclear chromosomes [36]. Because nuclear copies of regular mitogenomes exist, their occurrence for swinger-transformed mitogenomes is plausible, and would consist in itself a possible independent confirmation of the existence of swinger sequences, as previously discussed [103]. In addition, the possibility of swinger transcription of regular numts in the nucleus can't be ruled out.
Previous analyses [103] showed that the majority of detected swinger reads have mitochondrial origins. On average, alignments between swinger-transformed mitogenomes and RNA seq reads have higher identity percentages than between the same swinger mitogenome sequences and nuclear chromosome sequences. This is the case for a statistically significant majority of comparisons of identity percentages obtained between RNA reads and the swinger-transformed mitogenome, versus that between the same swinger-transformed mitogenome region and nuclear chromosome sequences. This suggests that most potential swinger numts diverge from their ancestral mitogenomic sequence, and that most RNA reads aligning with swinger-transformed mitogenomes have mitochondrial origins, because the swinger-transformed mitogenomes resemble on average more RNA reads than putative swinger numt(s) [103]. The ‘Discussion’ below develops these points in relation to potential nuclear origins of chimeric peptides.
1.6. Chimeric RNAs due to fusion between different RNAs
The term ‘chimeric’ transcripts has been used in the literature for a different type of RNA than the contiguous regular- and swinger-transcribed RNAs [101]. These other types of chimeric RNAs refer to two or more different transcripts produced each by regular polymerization, on the template of disjunct DNA regions. These RNAs are then fused by natural [60], [132] or artificial reverse-transcription-associated phenomena [131]. These chimeric RNAs differ from the regular-swinger RNAs in the sense that for the latter the transcription process is chimeric (part regular-, part swinger transcriptions), but not in terms of their templating DNA regions, which are contiguous, not disjunct. It is possible that some unknown sequencing artifacts produce some of the detected swinger reads, but the non-random mapping of detected swinger peptides on detected swinger RNA reads, as previously described [103] shows that most swinger reads exist while translation occurs in the cell, and hence are not artifacts.
1.7. Swinger polymerization creates new genomic sequences
Another type of analyses detected swinger repeats within the regular mitogenome. Swinger repeats are usually short repeats that can only be detected when taking into account swinger transformations. These short sequences are inserted within the regular mitogenome, suggesting that natural retrotransposition of swinger RNAs produces novel DNA sequences [101]. They are more frequent and longer than expected by chance, and their length is proportional to the probability that the specific swinger transformation conserves circular code signals that presumably maintain the ribosomal translation frame in the gene. The natural circular code is a punctuation code within the genetic code consisting of 20 codons that as a group, have properties that enable protein coding frame retrieval [4], [22], [55], [56], [57]. This indicates that insertion of swinger sequences in the human mitogenome depends on their capacity to integrate protein coding genes without disrupting punctuation that presumably enables ribosomal detection of the coding frame.
1.8. Chimeric peptides
Recent analyses show convergent frequencies between swinger RNAs sequenced by classical and next generation (RNAseq) sequencing methods [103]. Hence swinger RNA occurrence is relatively well confirmed by data from independent methods and research teams. Here analyses complement at peptide levels results on chimeric transcripts. The existence of transcripts that are part regular (untransformed), part swinger RNAs, with an abrupt switch between these parts, predicts the existence of ‘chimeric’ peptides matching translation of such chimeric transcripts. Hence MS/MS mass spectra of peptide data (from [33]) previously used to detect swinger peptides [103] are reanalyzed here, using the same methods as by Seligmann [103]), searching for peptides matching in part the translation of the untransformed human mitogenome, and in part the translation of the swinger-transformed, contiguous mitogenome sequence. Chimeric peptides are peptides where swinger-encoded parts of a peptide are contiguous with parts translated from regular RNA. These would be stronger evidence for translation of swinger RNA than previous detections of entirely swinger-encoded peptides because the regular encoded parts function as matched positive controls, directly associated with swinger-encoded parts. In addition, chimeric peptides could suggest that swinger peptides are integrated within otherwise regular proteins, a further small step to understand functions associated with swinger phenomena.
2. Materials and methods
The revised Cambridge reference sequence for the human mitochondrial genome (NC_012920, [3]) was cut according to a running window of 270 nucleotides. Analyzes do not account for known mitochondrial polymorphisms, as this would expand analyzes beyond computing powers. The six frames of each of these nucleotide sequences of 270 bases were translated into the corresponding hypothetical peptides according to the vertebrate mitochondrial genetic code, after the 90 nucleotide-long mid-third of that sequence was swinger transformed according to each of the 23 swinger transformations. Hence each running window (around 16,300 in total) is represented by the six peptides translated from each of its 23 partly swinger-transformed versions (6 frames × 23 swinger versions = 138 hypothetical chimeric peptides for each of the 16,300 running windows). The window length of 90 codons/amino acids is designed to match the length of the longest (non-chimeric) peptides (up to 40 amino acids, [103]) previously detected in this dataset [33]. All translated hypothetical peptides are used by Thermo Protein Discoverer to predict a theoretical mass spectrometry distribution, which is matched with observed MS/MS mass spectrometry data from Gueugneau et al. [33]).
Stops are translated as ‘X’, which Thermo Proteome Discoverer considers by default as leucine/isoleucine (these have equal masses, and are indistinguishable by mass spectrometry). Peptides including stops are duplicated 18 times, replacing ‘X’ by one of the 18 remaining amino acid species, excluding leucine and isoleucine. Hence predicted peptides include the possibility that any amino acid could be inserted at stops. Analyses assume that all stops in a single predicted peptide are translated by the same amino acid. Hence the 138 peptides for a single window of 270 nucleotides, if it includes at least one stop (the majority of cases), are represented 19 times, inserting X and each of the remaining amino acids at stops (19 x 138 = 2622 chimeric peptides). In total, approximately 42.7 million hypothetical chimeric peptides were tested.
Consensus searches were handled with the Sequest (Thermo Fisher Scientific, Illkirch) algorithm with the following mass tolerances: Parent = 1 Da and Fragment = 0.5 Da (monoisotopic masses). Fixed carbamidomethyl (C) and variable Oxydation (M) modifications were activated, as well as the lysine → pyrrolysine modification, and only one missed trypsin cleavage was allowed. False discovery rate was estimated against a reverse decoy database using the Percolator algorithm. No protein grouping was allowed since the database only contained non redundant entries. Peptides with false discovery rate q < 0.05 and score Xcorr > 1.99 were considered identified. The score Xcorr is a likelihood of match between expected and observed MS/MS data that is unaffected by peptide length. Further explanations on peptide detection and characterization by the software are given in the Discussion. Observed mass spectra were compared separately to predicted peptides 19 times, each time inserting a different amino acid at stops. Here analyzes test the existence of a specific group of peptides, namely chimeric peptides. The false discovery rate q is adapted to such populations of detected items [41]. Results also indicate the posterior error probability PEP, an estimate of detection error specific to each individual peptide, which might be useful in the future, when analyzes focus on specific peptides, rather than on a population of peptides. Results are not analyzed according to this criterion more adapted to studies focusing on specific individual peptides.
3. Results
3.1. Chimeric peptides
Analyses detect according to the filtering criteria 1301 chimeric peptides, among approximately 42.7 million chimeric peptides produced by combinations of stop codon-amino acid insertions, swinger transformations and frames for the running window of 270 nucleotides (illustration in Fig. 1). Hence chimeric peptides are detected for approximately 3 among 100,000 hypothetical chimeric peptides. This is 200 times less than the rate of detection for ‘regular’ non-chimeric swinger peptides, using the same criteria and the same data, approximately 6 per 1000 predicted swinger peptides [103].
Previously detected chimeric human mitochondrial RNAs are about 3% of all RNAs detected with at least some swinger part. Part of the discrepancy between chimeric RNA versus peptide detections probably results from the fact that proteomic analyses only considered abrupt switches between regular and swinger parts of peptides. Blast analyses detecting RNAs are not limited by this consideration, and can detect RNAs where the switch is not abrupt: in a transition sequence between regular and swinger transformed sequences, nucleotides seem random. Hence for practical reasons, detection of chimeric RNAs encompasses more possibilities than chimeric peptide detection, explaining lower rates of chimeric peptide detection (relative to rates of swinger peptide detection); these are lower than rates of chimeric RNA detection (relative to swinger RNA detection rates).
Here we focus specifically on chimeric peptides for which each regular and swinger parts have more than 8 amino acids. This is because considering 19 different amino acid species (merging leucine and isoleucine), the e value for 42.7 million potential chimeric peptides is about 0.0001 for amino acid sequences of 9 residues (42,000,000 × 1/19− 9). Hence the match of each regular and swinger part of the detected peptide with the predicted chimeric peptide is unlikely to be due to chance, as estimated by this approximate e value.
This restricts the sample of 1301 detected chimeric peptides to 186 chimeric peptides of at least 18 residues, from various swinger transformations and stop-amino acid insertions (Table 1). Among these 186 chimeric peptides, the regular-encoded part of the peptide corresponds to the 5′ part of the peptide for 41% of the 186 chimeric peptides. This means that a statistically significant majority of chimeric peptides (two tailed sign test, P = 0.0061) correspond to the 5′ translation of swinger RNA and 3′ translation of regular RNA. Note that this statistically significant bias could not occur if detected chimeric peptides were due to random detection artifacts, strengthening the suspicion that results reflect a biological reality. Hence 41% of chimeric peptides reflect translation of regular transcripts that switch at a given point to swinger transcription. Frequencies and mean lengths of chimeric peptides, for each swinger type (Table 2) show that the regular (non-swinger) part of the chimeric peptides is on average slightly longer than the swinger part, though this difference is not statistically significant.
Table 2.
Frequencies and lengths of chimeric swinger peptides detected in Table 1: Columns indicate: swinger type; peptide number; mean PSMs; mean amino acid number in peptide non-swinger part; mean amino acid number in peptide swinger part.
| Swinger type | N | PSMs | Reg | Swinger |
|---|---|---|---|---|
| A ↔ C | 3 | 22.0 | 17.0 | 10.3 |
| A ↔ G | 4 | 96.0 | 14.8 | 14.0 |
| A ↔ T | 5 | 209.6 | 11.6 | 10.0 |
| C ↔ G | 4 | 139.0 | 11.3 | 14.0 |
| C ↔ T | 7 | 225.1 | 14.7 | 11.7 |
| G ↔ T | 1 | 1.0 | 16.0 | 11.0 |
| A ↔ C + G ↔ T | 11 | 59.6 | 13.8 | 14.2 |
| A ↔ G + C ↔ T | 10 | 103.0 | 13.2 | 14.4 |
| A ↔ T + C ↔ G | 12 | 31.2 | 12.3 | 14.8 |
| A → C → G → A | 10 | 52.2 | 14.6 | 13.7 |
| A → C → T → A | 5 | 47.0 | 14.2 | 13.6 |
| A → G → C → A | 9 | 91. 9 | 14.1 | 10.2 |
| A → G → T → A | 8 | 33.3 | 18.3 | 12.4 |
| A → T → C → A | 7 | 230.3 | 12.1 | 14.7 |
| A → T → G → A | 9 | 18. 8 | 18.3 | 12. 8 |
| C → G → T → C | 8 | 52.4 | 18.4 | 12.9 |
| C → T → G → C | 5 | 179.2 | 11.4 | 12.6 |
| A → C → G → T → A | 10 | 82.1 | 14.0 | 14.3 |
| A → C → T → G → A | 14 | 71.8 | 13.6 | 11.9 |
| A → G → C → T → A | 5 | 268.4 | 13.2 | 11.4 |
| A → G → T → C → A | 7 | 173.1 | 12.1 | 13.6 |
| A → T → C → G → A | 18 | 33.7 | 14.6 | 12.3 |
| A → T → G → C → A | 13 | 92.6 | 11.8 | 12.7 |
3.2. Swinger peptides and chimeric peptides
Note that chimeric peptides, due to their part that matches translation of regular transcripts, differ in mass spectrometry properties from peptides entirely translated from swinger RNA, even if these have the exact same swinger sequence. Hence detection of chimeric peptides with swinger parts overlapping previously detected ‘regular’ swinger peptides (as the swinger peptides described by [103]) would be strong, independent methodological confirmation that positive results are not artifacts. Indeed, the swinger parts of eight chimeric peptides in Table 1 overlap with one of the 263 previously described swinger peptides [103]. These previously described swinger peptides cover on average 1.1% of the swinger-transformed mitogenome, expecting approximately 2 overlaps with chimeric peptides in Table 1 if no association exists between the two independent analyses. This means that chimeric peptides map on previously described swinger peptides 4 times more frequently than expected. This association between two independent searches confirms that results are not false positive matches between the mass spectrometry data and some predicted hypothetical chimeric peptides among a very large number of predicted hypothetical chimeric peptides. In addition, note that even if detected swinger and chimeric peptides correspond to the same swinger region, the corresponding MS/MS mass spectra differ, because for chimeric peptides mass spectra include also the adjacent residues translated from regular, untransformed RNA, while for swinger peptides, mass spectra do not include the latter residues. This non-random correspondence between swinger peptides and swinger parts of chimeric peptides suggests that translation of swinger RNAs is not random, and probably specific to some mitogenome regions.
3.3. Swinger RNA and chimeric peptides
Previously detected swinger peptides preferentially map on human mitogenome regions covered by independently detected swinger RNAs [103]. Their numbers increase with numbers of detected swinger transcripts. These positive associations between swinger RNA and swinger peptides can also be expected for chimeric peptides described in Table 1, Table 2. Such associations would confirm that the detected chimeric peptides actually exist, because they would match two independent material evidences, peptides, and RNA fragments.
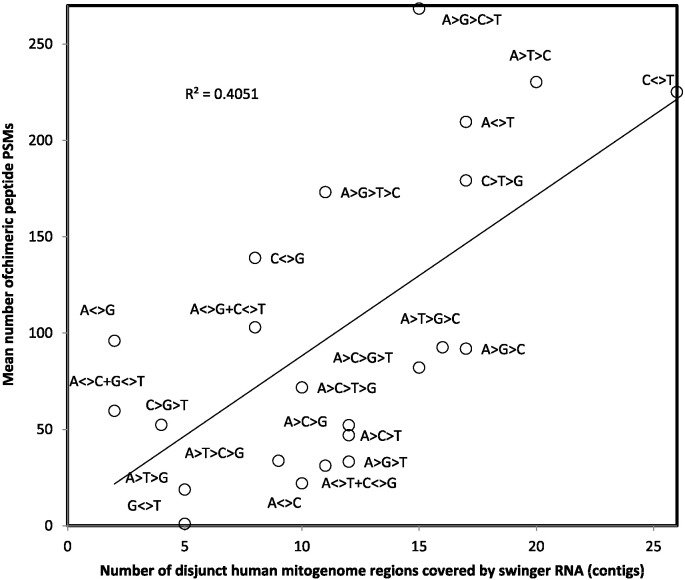
The mean number of PSMs (peptide spectrum matches) for chimeric peptides increases as a function of the number of human mitogenome regions covered by swinger RNA (also called contigs), for the swinger type corresponding to the swinger part of the chimeric peptides (Fig. 2). Swinger transcriptomic data are from Seligmann [103]). Chimeric peptides presumably reflect translation of chimeric RNAs, along part regular, and part swinger transcription rules. Hence amounts of chimeric peptides should reflect numbers of possible transitions between regular and swinger RNAs, estimated by the number of swinger contigs previously described by Seligmann [103]). Indeed, a positive association between PSMs of chimeric peptides and swinger RNA contigs exists (r = 0.64, one tailed P = 0.0006), strengthening confidence in the validity of results, and corresponding with previous results for swinger peptides [103]. Note that similar correlation analyses for numbers (not PSMs) of detected chimeric peptides do not yield statistically significant associations with contig numbers.
Fig. 2.
Mean number of PSMs detected for chimeric peptides, as a function of the number of disjunct human mitogenome regions covered by swinger RNA (RNA data from [102]). The positive association indicates the expected causal link between swinger RNA and chimeric peptides.
The swinger part of 8 chimeric peptides (marked by * in Table 1) maps on human mitogenome regions also covered by the adequate type of swinger RNA (six swinger types, two matches for A → G → T → A and A → C → G → T → A, and one match for each A → G → C → T → A, A → G → T → C → A, A → T → C → G → A, and A → T → G → C → A swinger transformations). Considering the overall mitogenome coverage by swinger RNAs (on average 2.6% of the genome), lack of association between swinger RNAs and the swinger part of chimeric peptides would expect 4.76 matches across all 23 swinger transformations, with 0.21 peptides for the average swinger transformation. This predicted number for specific swinger transformations was always < 0.5 peptides. Detecting at least one match for six among 23 swinger types, when less than 0.5 are expected for all 23 swinger transformations has P = 0.022 according to a two-tailed Fisher exact test. This indicates that chimeric peptides associate with detected swinger RNA, though this association is weaker than the previously described association between swinger RNA and swinger peptides [103].
3.4. Chimeric peptides: strong validation of swinger sequences
Chimeric peptides are in terms of confirmation of swinger polymerization only secondary evidence, because peptides are translated from RNA, as compared to previous descriptions of swinger RNAs and chimeric swinger RNAs [100], which directly result from swinger polymerization. This point is also valid for swinger peptides. However, detection of (numerous) peptides matching translation of contiguous parts of the mitogenome, where one part reflects regular transcription, and the other swinger transcription, is a strong methodological confirmation for swinger phenomena and associated translation into peptides, which is not implied by the detection of ‘pure’ swinger peptides. This is because the non-swinger part of the peptide is a positive control paired to its contiguous swinger part. Hence in addition of describing a further aspect of the biological phenomenon of swinger polymerizations, chimeric peptides are also a further validation of the phenomenon's existence.
3.5. Chimeric peptides integrated in regular proteins?
An important question associated to swinger sequences is their function: among others, do they code for functional proteins, and are swinger peptides integrated into regular, perhaps functional proteins? A reanalysis of Table 1 yields a first insight into these important questions. The regular (non-swinger) part of eleven peptides matches the sequence of six among thirteen known, regular, mitogenome-encoded proteins. Their swinger parts correspond to the translation of the contiguous swinger transformation of these genes, along nine (four symmetric, and five asymmetric) systematic nucleotide exchange rules (Table 3). Note that up to three chimeric peptides are detected for two large mitochondrial proteins (cytochrome c oxidase I and NADH:ubiquinone oxidoreductase subunit 5). It is plausible that such peptides are integrated within complete proteins. These sequence alterations could modulate (or not) the regular function of the protein, and not necessarily impair function.
Table 3.
Chimeric peptides from Table 1 with regular part matching proteins translated from known mitogenome-encoded genes. Swinger parts are underlined, gene identity is followed by the position of the ‘normal’ part of the peptide matching the regular translation of the gene in the regular protein. The swinger transformation and the amino acid inserted at stop(s) are also indicated. Peptide parts matching translation according to both nuclear and mitochondrial genetic codes are highlighted: peptide 100 could be translated in the cytosol on the base of RNA transcribed from mitochondrial inserts in the nuclear chromosome (numts), all remaining peptides could not, as at least on part of the peptide is incompatible with translation according to the nuclear genetic code. Analyses (see text) show that there are fewer detected peptides compatible with the nuclear genetic code than expected, and more than expected peptides compatible only with the mitochondrial genetic code.
| Table 1 # | Peptide | Gene | Position | Swinger rule | Stop |
|---|---|---|---|---|---|
| 100 | GASFLFIWNSLYLLFGAWAGVLGTALSLLIRAELGQPGNL | COX1 | 18–47 | A > T > G | r |
| 27 | SGWVEWSRHSVLLLLSLPVLAAGITMLLTDR | COX1 | 205–213 | A ↔ C + G ↔ T | s |
| 181 | LLKECLSLASVPATPPYHTFEEPVYMK | COX1 | 500–512 | A > T > G > C | x |
| 136 | NQMIQALLITILLGLYFTLLSIVTAGTVFGLR | COX3 | 157–168 | A > C > T > G | e |
| 169 | AASHPVPVPMTLLMLGLLTNTLTMYQWWR | COX3 | 41–59 | A > T > C > G | a |
| 23 | DVSGPSSPSSSLMTLTLFSPDLLGDPDNYTLANPLNTPPY | Cyt B | 238–267 | C ↔ T | y |
| 19 | NPSLSISVPSTRHVSMPITISSIPPQTTEMCLMK | ND1 | 305–318 | C ↔ T | t |
| 38 | WALFLSGTDSSSVSLAPLAATGSWGGLNQTQLR | ND2 | 165–176 | A ↔ G + C ↔ T | n |
| 34 | SLKQNWDFSFNSSTMVVAGIFLLIR | ND5 | 249–262 | A ↔ C + G ↔ T | f |
| 80 | IVAFSTSSQLGLMVLEVPVGVK | ND5 | 301–313 | A > G > T > C | d |
| 7 | LLGAVPLASASLTIGSLALAGMPFLTGFYSKDHIIETANMS | ND5 | 374–402 | A ↔ G |
These 11 chimeric peptides integrated in regular proteins represent 5.9% of all 186 detected chimeric peptides. Considering that regular mitochondrion-encoded proteins have a total length of 3789 amino acids, the regular proteins represent 11.43% of the total number of amino acids that could be translated from the positive and negative strands of the human mitogenome. This means that chimeric peptides embedded within regular coding sequences are half as frequent as expected (5.9 versus 11.43%). This principle is further strengthened when examining the number of PSMs (number of identified peptide spectra matching a hypothetical peptide) for these 11 regular-protein-integrated chimeric peptides, as compared to the mean number of PSMs for all chimeric peptides detected for that swinger transformation: their PSMs is in all but one case (peptide 80 in Table 1) lower than the mean PSMs of other chimeric peptides for that swinger transformation. Hence chimeric peptides within regular proteins are rarer, and less expressed (as far as PSMs numbers can be trusted to reflect peptide abundances), than chimeric peptides translated from non-coding sequences, and non-coding frames of regular protein coding genes.
3.6. The natural circular code and swinger RNA, peptides and chimeric peptides
An anonymous reviewer suggested examining whether properties of chimeric peptides can be predicted from frameshift error-correcting properties of the natural circular code. Indeed, abundances of detected swinger RNAs in GenBank's EST database are proportional to reading frame retrieval (RFR) after swinger transformation of the natural circular code [58]. In this context, RFR, which estimates the capacity of the natural circular code to retrieve the protein coding frame, is calculated for the 20 codons that form the natural circular code, after each of the 23 swinger transformations: some codons belonging to the natural circular code are transformed into another codon included in the natural circular code, meaning that this property is invariant in relation to that codon and swinger transformation. RFR estimates this across all 20 codons of the natural circular code, for each swinger transformation. The length of swinger repeats in the human mitogenome is proportional to the RFR of the swinger transformation [101], which suggests that RFR affects insertion rates of swinger repeats in protein coding regions, and hence could also affect chimeric peptide production.
The association between RFR and swinger RNA abundances for EST sequences occurs also for mitogenome coverage by swinger RNA reads sequenced by RNAseq in the transcriptome by Garzon et al. [30]) (Pearson correlation coefficient r = 0.528, one tailed P = 0.005). For swinger peptides as described by Seligmann [103]), the mean number of PSMs also increases with RFR (r = 0.364, one tailed P = 0.044). This positive association between mean PSMs numbers and RFR is also detected for chimeric peptides from Table 1 (r = 0.367, one tailed P = 0.043). These two results are independent, also because mean PSMs of swinger and chimeric peptides are only weakly correlated (r = 0.24, P > 0.05). Hence detections of chimeric and swinger peptides are proportional to extents by which swinger transformations conserve natural circular code ‘frame’ punctuations. Note that RFR, as mitogenome contig numbers in a previous section, associate with mean PSMs, rather than numbers of detected peptides, suggesting that in the context of these specific data, PSMs are better quantitative estimates than other variables.
4. Discussion
4.1. Statistical validity of peptide detections by mass spectrometry
An anonymous reviewer of a previous version indicated that detection of peptides with masses approximately matching the numerous possibilities produced by translation of all potential chimeric RNAs could be due to chance, due mainly to the large number of hypothetical chimeric peptides. Indeed, considering all 19 possible amino acids inserted at stops introduces a ‘fudge’ factor that enables adapting many hypothetical peptides to an actual fragment with a similar mass. Note that 28 among 186 (15%) detected peptides lack stops, invalidating this argument for several detected chimeric peptides. Independently of this, there are three reasons why this important point does not invalid the remaining results on chimeric peptides presented here. This is first because mean chimeric peptide PSMs converge with corresponding swinger RNA contig numbers, an independent type of data unrelated to the problems of proteomic analyses, already discussed above.
The other two points relate to the nature of the MS/MS mass spectrometry analyses themselves. The factor ‘detection by chance’ is integrated into the detection software used by Thermo Proteome Discoverer. The software compares the match between the mass spectrum of the actual fragment and the predicted mass spectrum of the hypothetical peptide, and its match with a dataset of decoy (false, negative controls) predicted peptides. The q value estimates the false detection rate (FDR, see explanations by [41]) of a peptide based on comparing matches by the actual predicted peptides and the decoy peptide database. This q is a probability of detection corrected for the false detection rates within the population of positive results (classical P values consider the whole population of statistical decisions, not only the subpopulation of positives). Hence the reported detections account for matches due to chance, considering the various parameters of the samples analyzed/compared, among them in particular sample sizes.
The third point relates to the nature of the statistic whose distribution is used to evaluate the above mentioned q (FDR). It is Xcorr, the cross correlation of the goodness of fit between the experimental peptide fragments and theoretical mass spectra. This integrates fits with each b and y ions, which correspond to asymmetry in the physical fragmentation of peptide bonds within the detected peptide, resulting into shorter peptide subparts: b ions occur when the residue's N-terminal is charged, y ions when the C-terminal is charged. Hence the match between the observed and the predicted peptide is not based solely on the similarity between their total masses, but also on fit between distributions of masses of sub-fragments of the (expected and observed) peptides, and this separately for b and y ions. The Xcorr statistic accounts, in addition to peptide size, for the number of matching masses of such sub-fragments. This allows inferring more precisely the residue sequences in the peptide, and means that peptide detection is not based only on a single measure, its total mass, but also on the mass of several subfragments.
In this context, the peptide ACD can function as a simplified example. Its mass corresponds to six possible peptides, ACD, ADC, CAD, CDA, DAC and DCA. Hence if ACD results from translation of swinger RNA, one can't assert that the observed mass is due to this peptide rather than any of the other five possibilities. However, Xcorr also considers the masses of subfragments of this peptide. Detection of a subfragment matching the mass of AC excludes four among the six possible peptides. A fragment matching the mass of CD matches only two peptides. If both subfragments AC and CD are detected, the characterization of the peptide ACD can be considered as assessed.
In addition, this process is done separately for b and y ions, because mass spectrometry analyses are in principle sufficiently precise to distinguish between these ions (remember that the precision of 0.5 Da of the analyzed data means a precision of half the mass of a hydrogen atom, which is also far less than the difference between amino acids with similar molecular masses). Hence Xcorr integrates information from both b and y ions, evaluating whether that information is congruent with the observed data. This procedure, coupled to q values based on comparisons of the Xcorr distribution obtained for negative controls (decoy peptides), renders detections relatively robust, despite fuzzy factors. In fact, large numbers of predicted peptides are necessary to estimate properly the distribution of random Xcorrs. The last point stresses that q (as P) values account for numbers of predicted peptides.
4.2. Confirmation of chimeric peptides by Waters technology
An anonymous reviewer suggested to confirm the existence of chimeric peptides by additional, independent mitochondrial proteomic data. In this context, I focused on another analysis of trypsinized human mitochondrial peptides [1], extracted by a more up to date MS/MS technology (Waters, Milford, MA, http://www.waters.com). This technique yields more accurate mass estimates than the method used by [33] (0.5 Da for the latter versus 5 ppm for the Waters method, hence about 10 × more accurate estimates).
Analyses of the twelve samples from Alberio et al. [1]) by the software PLGS yield relatively few hits matching chimeric peptides considering only peptides where each regular and swinger-encoded parts are each at least nine amino acids long. One peptide matches significantly according to PLGS a chimeric peptide whose swinger part (underlined) matches swinger transformation A → T → G → A, LVSASVEMNQQQVPGSAGR (the regular part are residues 4228–4237 translated from the third frame of the negative strand of the human mitogenome). The other peptide detected in these data has a swinger part that matches transformation C → G → T → C, SAAAARAGSACCLTSTAVTDRNLNTTF, the regular-encoded part corresponds to COX1, residues 211–219 in that regular mitochondrion-encoded protein.
Hence a different technology detects within independent mitoproteomic data peptides matching translations of chimeric RNAs, with one part regular, the other swinger transformed RNA. Hence, at least qualitatively, these independent data and technology confirm the existence of chimeric peptides and their integration in regular mitochondrial proteins. A more detailed description of ‘regular’ swinger peptides (meaning peptides entirely coded by swinger transformations of the mitogenome (unlike chimeric peptides that are in part regular-, in part swinger-encoded)) detected in the data from Alberio et al. [1]) will be presented elsewhere.
These results from data by Alberio et al. [1]) are too scarce to indicate whether chimeric peptides are produced according to a non-random profile. However, the non-random convergence between chimeric and entirely swinger peptides (detected in the same dataset from [33]) noted in a previous section in Results is in itself an indication that swinger-encoded peptides or parts of peptides are non-randomly produced.
4.3. Nuclear mitogenome copies
Previous transcriptomic analyses that detected non-canonical RNAs transformed according to systematic rules, such as deletions of mono- and dinucleotides after each transcribed trinucleotide (producing delRNAs, [102]), and swinger transformations [103], included controls that account whether the transformed mitogenome versions match nuclear chromosome sequences: mitogenome analyses are frequently contaminated by such chromosomic pseudogenes [9], [10], [48], [49], [50], [51], [62], [66], [67], [123], [124], [125], [133], [134].
These previous analyses blasted the swinger-transformed mitogenome versions versus the (regular) human nuclear chromosomes. For transformed mitogenome regions aligning with both transcriptomic reads and chromosomes, similarities between the transformed mitogenome and the RNA contigs were compared with the corresponding similarities between the same transformed mitogenome region and the chromosomes. For each del- and swinger RNAs (non-canonical RNAs), similarities with RNA contigs were greater than those with chromosome sequences in significant majorities of cases [102], [103], as already discussed above for swinger RNA reads.
These results indicate two major issues. First, overall, RNA contigs result from non-canonical transcriptions of the mitogenome, the point that was being tested. Second, the observation that chromosome sequences match transformed versions of the mitogenome suggests that chromosomes include inserts of mitogenomic origins that were transformed according to systematic rules. The observation that these are on average less similar to the transformed mitogenome than RNA contigs suggests that these transformed mitochondrial sequences inserted in nuclear chromosomes mutated apart from the original sequence, as expected for inserts lacking function in the cell's nucleus [16], [24], [29], [34], [35], [36], [38], [40], [52], [54], [63], [65], [71], [77], [113], [116], [117], [118], [119], [126], [128], [129].
4.4. Peptides translated according to nuclear or vertebrate mitochondrial genetic codes
Similar-minded analyses at the peptide level can test whether chimeric peptides in Table 1 were translated according to the human mitochondrial or the nuclear genetic codes. For that purpose, the regular and swinger transformed versions of the human mitogenome were translated according to the standard genetic code, which differs from the vertebrate mitochondrial genetic code by the reassignment of codon ATA from Met to Ile, of TGA from Trp to stop, and AGR from stop to Arg [23]. These four codons are 6.25% of all 64 codons.
Each swinger- and regular-encoded part of detected chimeric peptides has at least 9 amino acids. Hence the probability of detecting chimeric peptides that would have identical sequence according to both genetic codes is (1 − 0.0625)− k, where k is the total length of the peptide. This principle is applied to the chimeric peptides in Table 1 so as to calculate the predicted number of peptides, for each size category, that is expected to match translation according to both nuclear and mitochondrial genetic codes. Lengths of chimeric peptides in Table 1 range from 18 to 42 residues, a total of 24 length categories. The observed number of chimeric peptides compatible with translation according to both genetic codes (in total 30 among the 186 chimeric peptides) is lower than expected in 16 among 24 size categories. Obtaining this result has P = 0.038 according to a one-tailed sign test. This means that, considering the length of chimeric peptides, there are statistically significantly fewer than expected peptides with sequences compatible with translation according to the nuclear genetic code.
The same principle can be applied to chimeric peptides in Table 1 whose sequences are only compatible with translation according to the mitochondrial genetic code, separately for each the regular- and the swinger-encoded parts. Here, the observed number (54) should be larger than the predicted number, if the sample is biased towards mitochondrion-encoded/translated peptides. Considering that 6.25% of codons differ in codon-amino acid assignments between the two genetic codes, the total expected number of chimeric peptides, considering their size, containing at least one of the 4 codons with coding assignment differing between nuclear and mitochondrial genetic codes is 35.97. This number is far lower than the observed 54 according to a chi-square test (P = 0.0027). Hence chimeric peptides with sequences compatible only with translation according to the mitochondrial genetic code are significantly more frequent than expected. This bias confirms the mitochondrial origin of chimeric peptides in Table 1. The number of peptide length categories where more observed peptides than expected are only compatible with mitochondrial translation is again 16 among 24 length categories, which has P = 0.038 according to a one tailed sign test.
These analyses show that detected peptides are more likely translated according to the mitochondrial genetic code than according to the nuclear genetic code. Note that translation, within the mitochondrion, according to the nuclear code is possible: it potentially depends for some codons upon the presence of cytosolic tRNAs, which could be occasionally imported in mitochondria [21], [32], [39], [44], [75], [76], [78], [112], [114]. However, this rationale is not symmetric: cytosolic translation according to the mitochondrial genetic code is much less probable than the opposite, so that nuclear origins are not compatible with the results obtained.
In fact, whether peptides have cytosolic or mitochondrial origins does not actually affect the main point that is addressed here, which is that these peptides were translated in part from swinger RNA. The same point applies to the potential nuclear (numt) origin of swinger-transformed mitochondrial DNA: independently of the location of the process, detection of chimeric peptides implies that swinger transformations occurred, whether during transcription of the regular mitogenome or nuclear inserts, or during numt insertion, possibly by natural swinger retrotranscription. This does not exclude the possibility that some detected chimeric peptides originate from the cytosol, but stresses the fact that most are mitochondrial, and that this issue is not directly relevant to the fact that swinger RNAs, chimeric RNAs, and corresponding peptides, exist, independently of the question of which cellular compartments produce them.
4.5. Few chimeric peptides in regular proteins translated from mitochondrion-encoded genes
Chimeric peptides in Table 3 have regular parts that match sequences of regular mitochondrial proteins encoded by mitochondrial genes. These are about 5% of all 186 detected chimeric peptides. Peptides translated from regular mitochondrial genes represent about 11% of the total length potentially translated from the complete mitogenome, considering all six frames. Hence these 11 chimeric peptides potentially integrated in regular mitochondrial proteins are half as frequent as one could expect. Their PSMs is lower than for other chimeric peptides. These are hence rarer and less expressed than one could expect. Possibly, chimeric peptides integrated in regular proteins perturb proper protein folding. Incorrect folding induces various degradation mechanisms associated with mitophagy [5], [42], [122], which could explain that only few chimeric peptides are detected within regular proteins. These findings are not incompatible with the possibility that at least some swinger transcripts and peptides are functional.
4.6. Secondary structure formation by swinger transformed RNA and swinger RNA detection
Secondary structure formation by self-hybridization of DNA/RNA groups bijective transformations into three classes of each eight transformations. These share self-hybridization properties within each class [27], [28]. This means that seven bijective transformations (including A ↔ T + C ↔ G) conserve self-hybridization properties of the original, untransformed sequence. Secondary structure formation by swinger RNA associates with swinger RNA detection [104], but these groupings/properties do not correlate with differences in chimeric or swinger peptide abundances/PSMs (not shown). The issue of regulation of alternative mitochondrial transcriptions, respectively post-transcriptional splicing, in relation to secondary structure formation by transformed RNA [61] remains unclear: a positive association exists between RNA occurrence and secondary structure formation for regular and swinger RNAs, but for transcripts resulting from systematic deletions (delRNAs), a negative association exists between secondary structure formation after deletions and delRNAs [105].
4.7. Swinger transformations, RNA–DNA differences (RDDs) and heteroplasmy
Specific non-random point differences occur between DNA and RNA sequences, either due to nucleotide substitutions [47] or inserts/deletions [15], including for human mitochondrial transcripts [8], [37], [59]. These RDDs appear shortly after transcripts exit polymerases [130], suggesting RDDs are due to post-transcriptional edition. The systematic repetition of transformations over long sequence stretches that characterize swinger RNA seem less likely produced by post-transcriptional edition than some unusual stabilized polymerase state, however, at this point, no possibility can be excluded, and potential connections of del- and swinger RNAs with RDDs should be kept in mind.
For the same reason, and by definition, punctual mitochondrial heteroplasmies [45], [74], [115], [121] could not account for swinger parts of chimeric peptides, because these have to be translated from sequences differing from standard mitogenome sequences by far more than punctual nucleotide substitutions. Mitochondrial length heteroplasmies are common (49% of individuals, [64]), and in principle could, by chance correspond to swinger-like inserts in the mitogenome. Considering the seven regions containing length heteroplasmies described by Ramos et al. [64]) (therein table 3), only three among 186 chimeric peptides in Table 1 (peptide numbers 3, 156 and 157) potentially overlap (and this only in part) with these length heteroplasmies. Hence length heteroplasmies map non-randomly on chimeric peptides (3 among seven). Hence some presumed chimeric peptides might be translated from regions presenting length heteroplasmies, but this explanation is compatible with, at most, a small minority of chimeric peptides. Hence heteroplasmy could not explain chimeric peptides.
4.8. Translation increasing codon size or transcription systematically deleting nucleotides
It is important to note in the broader context of the discussion of results that further little known mechanisms increase the coding potential of sequences. A different, sometimes tRNA-based mechanism produces an alternative decoding of sequences, that of systematic frameshifting, which expands the codon from three to four (or five) nucleotides [6], called here tetracodons or pentacodons. This could result from systematic ribosomal slippages, a phenomenon that would correspond to programmed frameshifts (e.g. [25], [43]), but occurring systematically, and serially; and/or from translational activities of tRNAs with expanded anticodons [68], [120], [127]. These cases relate to previously described isolated frameshift mutations, interpreted as isolated tetra-, pentacodons.
The hypothesis of an early genetic code based on quadruplets was suggested by Baranov et al. [6]) to solve the problem that the weak triplet codon-anticodon interactions could not occur from a thermodynamic point of view in the absence of ribosomes, especially if these occurred at high temperatures [17], [18], [19], [20]. Molecules as complex as ribosomes probably were absent at proto-life stages. Codon-anticodon interactions between four (or more) base pairs are more stable than those between three base pairs. Symmetry considerations also enable the deduction that the primeval genetic code was based on a subset of 64 quadruplets, called the tesserae, specifically for the vertebrate mitochondrial genetic code [31].
The expanded codon hypothesis is that modern genes include overlapping coding regions that consist of series of tetra- or pentacodons. This hypothesis is compatible with bioinformatic analyses where all eight frames of mitochondrial genes were translated assuming tetracodons. Blast analyses detected alignments between parts of these hypothetical tetracoded peptides and regular proteins in GenBank. Several other analyses based on codon usages in these tetracoding sequences confirm their special coding status, including higher GC contents than in non-tetracoding neighboring mitochondrial sequences. This corresponds to the prediction that tetracoding is an adaptation to translation at high temperature [89]. This point was further confirmed by a positive correlation between predicted tetracoding in lizard mitogenomes and mean body temperature in these lizard species [109]. Accordingly, overlap coding by tetracodons increases with temperature.
At this point, and besides the proven existence of decoding mechanisms for isolated tetracodons, the strongest further evidence for the existence of protein coding regions based on tetracodons is the coevolution between predicted tetracoding regions and the predicted antisense mitochondrial tRNAs with expanded anticodons, which is observed in mammal and Drosophila mitochondria [88], [90], [95]. In addition, mitochondrial peptides matching translation of regular and swinger RNAs according to tetra- and pentacodons have been detected [103], as well as translation of delRNAs (or dRNAs), RNAs transcribed while systematically deleting every fourth, or every fourth and fifth nucleotide. Peptide translation of such transcripts uses regular tRNAs but produces peptides identical to those resulting from decoding by tRNAs with expanded anticodons of regular transcripts [102]. These delRNAs are produced by systematic deletions, every third nucleotide, and correspond at deletion level, to systematic nucleotide substitutions/exchanges. This predicts that chimeric peptides consisting in part of regular-translated, and in part tetra- or pentacoded peptides, might exist.
The strongest evidence for swinger-encoding is the association between detected swinger RNAs and detected swinger peptides. Analyses detecting mass spectra matching predictions according to translations of tetra- and pentacodons suffer the caveat that evidence is based solely on mass spectra, with the above discussed difficulties in asserting the robustness of results based only on proteomics. However, further analyses detected peptides matching translations, according to expanded codons, of swinger-transformed sequences, and showed their association with detected swinger RNA [103]. Hence from a methodological point of view, translation according to expanded codons of swinger RNAs is stronger evidence for tetra- and pentacoding than such translation of regular RNA because it is confirmed by the independent detections of two ‘unusual’ types of molecules, swinger RNA and corresponding peptides matching expanded codons.
4.9. Robustness of experimental design
An anonymous reviewer indicated that analyzes comparing transcriptome and proteome make sense only if data originate from individuals with the same phenotypes, and if possible the same tissues and even the same individual(s), however analyzes compare tumor transcriptome [30] with normal proteome [33]. This setup is indeed suboptimal. However, considering this point, RNA and peptide data converge (also in previous analyzes, [102], [103]) despite that RNA and peptide data originate from different tissues/individuals/phenotypes. This indicates that the phenomenon is general, and robust. This should not be surprising, because analyzes consider only RNA and peptides corresponding to the mitogenome. Most tissue-specific differences in mitochondrial RNA and protein profiles relate to molecules imported from the cytosol [13].
Methods used to detect the various types of unusual peptides take into account the large numbers of possibilities in matching observed and hypothetical mass spectra, so that positive detections are robust, and could not be due to chance.
Beyond methodological issues, occurrence of peptides coded by combinations of presumably unusual coding systems (translation of stops, together with translation according to expanded codons, and this for swinger RNAs), suggests that these basically ignored mechanisms expand more frequently than presumed the coding potential of genes, at least of the short mitogenomes. Detections of chimeric peptides, consisting of peptide parts corresponding to regular translation, adjacent to peptide parts matching translation of contiguous swinger RNA, strengthen confidence in the validity of results as positive controls, and expand our understanding of the phenomenon: swinger peptides are occasionally integrated in regular mitochondrion-encoded proteins, but their occurrence is downregulated.
5. Conclusions
-
1.
Analyses of MS/MS mass spectrometry data detect peptides matching the translation of chimeric transcripts, RNA following in part regular, and in part swinger-transformed transcription, assuming abrupt switches between regular and swinger transformed parts of the RNA.
-
2.
The 186 detected chimeric peptides (peptides consisting of a part encoded by regular RNA and a contiguous part encoded by swinger RNA) represent 3/100,000 among potential chimeric peptides, about 200 times fewer (6/1000) than detected swinger peptides (peptides entirely encoded by swinger RNA) in the same data. Eleven among these 186 chimeric peptides have a regular-encoded part that corresponds to proteins translated from classical mitochondrion-encoded genes.
-
3.
Chimeric peptides map on previously detected swinger RNA. This association is weaker than a previously described association between ‘regular’ swinger peptides and swinger RNAs [103].
-
4.
The vertebrate mitochondrial genetic code differs from the nuclear genetic code for four codons. Numbers of detected chimeric peptides that could be translated from human mitogenome sequences according to the nuclear genetic code are significantly fewer than expected considering the differences between the two genetic codes. This means that the majority of detected chimeric peptides are not cytosolic contaminations and were translated in the mitochondrion.
-
5.
Previous detections of swinger peptides (predicted products of translation of swinger RNA) suggested that swinger transformed RNA is translation-competent [103]. Chimeric peptides where the regular part corresponds to known mitochondrion-encoded proteins might be incorporated into the respiratory chain complexes. Chimeric and swinger peptides might affect known mitochondrial functions despite low abundances if they have regulatory functions. Results are compatible with the possibility that some proteins are encoded by swinger transformations, with yet unknown functions.
Acknowledgments
This work has been carried out thanks to the support of the A*MIDEX project (no ANR-11-IDEX-0001-02) funded by the « Investissements d'Avenir » French Government program, managed by the French National Research Agency (ANR). I thank Nicolas Armstrong for coaching the mass spectrometry analyses, and anonymous reviewers for constructive comments.
References
- 1.Alberio T., Bondi H., Colombo F., Alloggio I., Pieroni L., Urbani A. Mitochondrial proteomics investigation of a cellular model of impaired dopamine homeostasis, an early step in Parkinson's disease pathogenesis. Mol BioSyst. 2014;10:1332–1344. doi: 10.1039/c3mb70611g. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S.F., Madden T.L., Schaeffer A.A., Zhang J., Zhang Z., Miller W. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrews R.M., Kubacka I., Chinnery P.F., Lightowlers R.N., Turnbull D.M., Howell N. Reanalaysis and revision of the Cambridge reference sequence for human mitochondrial DNA. Nat Genet. 1999;23:147. doi: 10.1038/13779. [DOI] [PubMed] [Google Scholar]
- 4.Arquès D.G., Michel C.J. A complementary circular code in the protein coding genes. J Theor Biol. 1996;182:45–58. doi: 10.1006/jtbi.1996.0142. [DOI] [PubMed] [Google Scholar]
- 5.Ashrafi G., Schwarz T.L. The pathways of mitophagy for quality control and clearance of mitochondria. Cell Death Differ. 2013;20:31–42. doi: 10.1038/cdd.2012.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Baranov P.V., Venin M., Provan G. Codon size reduction as the origin of the triplet genetic code. PLoS One. 2009;4:e5708. doi: 10.1371/journal.pone.0005708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barthélémy R.-X., Seligmann H. Cryptic tRNAs in chaetognath mitochondrial genomes. Comput Biol Chem. 2016;62:119–132. doi: 10.1016/j.compbiolchem.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 8.Bar-Yaacov D., Levin A.G., Richards A.L., Hachen N., Rebolledo Jaramillo B., Nekrutenko A. RNA–DNA differences in human mitochondria restore ancestral form of 16S ribosomal RNA. Genome Res. 2013;23:1789–1796. doi: 10.1101/gr.161265.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bensasson D., Zhang D., Hartl D.L., Hewitt G.M. Mitochondrial pseudogenes: evolution's misplaces witnesses. Trends Ecol Evol. 2001;16:314–321. doi: 10.1016/s0169-5347(01)02151-6. [DOI] [PubMed] [Google Scholar]
- 10.Bensasson D., Feldman M.W., Petrov D.A. Rates of DNA duplication and mitochondrial DNA insertion in the human genome. J Mol Evol. 2003;57:343–354. doi: 10.1007/s00239-003-2485-7. [DOI] [PubMed] [Google Scholar]
- 11.Beznosková P., Gunišová S., Valášek L.S. Rules of UGA-N decoding by near-cognate tRNAs and analysis of readthrough on short uORFs in yeast. RNA. 2016;22:456–466. doi: 10.1261/rna.054452.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Breton S., Milani L., Ghiselli F., Guerra D., Stewart D., Passamonti M. A resourceful genome: updating the functional repertoire and evolutionary role of animal mitochondrial DNAs. Trends Genet. 2014;30:555–564. doi: 10.1016/j.tig.2014.09.002. [DOI] [PubMed] [Google Scholar]
- 13.Calvo S.E., Clauser K.R., Mootha V.K. MitoCarta2.0 : an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016;44:D1251–D1257. doi: 10.1093/nar/gkv1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Capt C., Passamonti M., Breton S. The human mitochondrial genome may encode for more than 13 proteins. Mitochondrial DNA. 2015 doi: 10.3109/19401736.2014.1003924. [DOI] [PubMed] [Google Scholar]
- 15.Chen C., Bundschuh R. Systematic investigation of insertional and deletional RNA–DNA differences in the human transcriptome. BMC Genomics. 2012;13:616. doi: 10.1186/1471-2164-13-616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dayama G., Emery S.B., Kidd J.M., Mills R.E. The genomic landscape of polymorphic human nuclear mitochondrial insertions. Nucleic Acids Res. 2014;42:12640–12649. doi: 10.1093/nar/gku1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Di Giulio M. The universal ancestor lived in a thermophilic or hyperthermophilic environment. J Theor Biol. 2000;203:203–213. doi: 10.1006/jtbi.2000.1086. [DOI] [PubMed] [Google Scholar]
- 18.Di Giulio M. The universal ancestor was a thermophile or a hyperthermophile. Gene. 2001;221:425–436. doi: 10.1006/jtbi.2003.3197. [DOI] [PubMed] [Google Scholar]
- 19.Di Giulio M. The universal ancestor and the ancestor of bacteria were hyperthermophiles. J Mol Evol. 2003;57:721–730. doi: 10.1007/s00239-003-2522-6. [DOI] [PubMed] [Google Scholar]
- 20.Di Giulio M. The universal ancestor was a thermophile or a hyperthermophile: tests and further evidence. J Theor Biol. 2003;221:425–436. doi: 10.1006/jtbi.2003.3197. [DOI] [PubMed] [Google Scholar]
- 21.Duchêne A.M., Pujol C., Maréchal-Drouard L. Import of tRNAs and aminoacyl-tRNA synthetases into mitochondria. Curr Genet. 2009;55:1–18. doi: 10.1007/s00294-008-0223-9. [DOI] [PubMed] [Google Scholar]
- 22.El Soufi K., Michel C.J. Circular code motifs near the ribosome decoding center. Comput Biol Chem. 2015;59A:158–176. doi: 10.1016/j.compbiolchem.2015.07.015. [DOI] [PubMed] [Google Scholar]
- 23.Elzanowski A., Ostell J. The genetic codes. NCBI. 2013. http://www.ncbi.nlm.nih.gov/Taxonomy/Utils/wprintgc.cgi?mode=t accessed 10V2016.
- 24.Ermakov O.A., Simonov E., Surin V.L., Titov S.V., Brandler O.V., Ivanova N.V. Implications of hybridization, NUMTs, and overlooked diversity for DNA barcoding of Eurasian ground squirrels. PLoS One. 2015;10:e0117201. doi: 10.1371/journal.pone.0117201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Farabaugh P.J. Programmed translational frameshifting. Microbiol Rev. 1996;60:103–134. doi: 10.1128/mr.60.1.103-134.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Faure E., Delaye L., Tribolo S., Levasseur A., Seligmann H., Barthélémy R.M. Probable presence of an ubiquitous cryptic mitochondrial gene on the antisense strand of the cytochrome oxidase I gene. Biol Direct. 2011;6:56. doi: 10.1186/1745-6150-6-56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fimmel E., Danielli A., Struengmann L. On dichotomic classes and bijections of the genetic code. J Theor Biol. 2013;336:221–230. doi: 10.1016/j.jtbi.2013.07.027. [DOI] [PubMed] [Google Scholar]
- 28.Fimmel E., Giannerini S., Gonzalez D.L., Struengmann L. Circular codes, symmetries and transformations. J Math Biol. 2015;70:1623-164. doi: 10.1007/s00285-014-0806-7. [DOI] [PubMed] [Google Scholar]
- 29.Françoso E., Gomes F., Arias M.C. A protocol for isolating insect mitochondrial genomes: a case study of NUMT in Melipona flavolineata (Hymenoptera: Apidae) Mitochondrial DNA A DNA MappSeq Anal. 2016;27:2401–2404. doi: 10.3109/19401736.2015.1028049. [DOI] [PubMed] [Google Scholar]
- 30.Garzon R., Volinia S., Papaioannou D., Nicolet D., Kohlschmidt J., Yan P.S. Expression and prognostic impact of lncRNAs in acute myeloid leukemia. Proc Natl Acad Sci U S A. 2014;111:18679-1868. doi: 10.1073/pnas.1422050112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gonzalez D.L., Giannerini S., Rosa R. On the origin of the mitochondrial genetic code: towards a unified mathematical framework for the management of genetic information. Nat Precedings. 2012 [Google Scholar]
- 32.Gowher A., Smirnov A., Tarassov I., Entelis N. Induced tRNA import into human mitochondria: implication of a host aminoacyl-tRNA-synthetase. PLoS One. 2013;8:e66228. doi: 10.1371/journal.pone.0066228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gueugneau M., Coudy-Gandilhon C., Gourbeyre O., Chambon C., Combaret L., Polge C. Proteomics of muscle chronological ageing in post-menopausal women. BMC Genomics. 2014;15:1165. doi: 10.1186/1471-2164-15-1165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hazkani-Covo E., Graur D. A comparative analysis of numt evolution in human and chimpanzee. Mol Biol Evol. 2007;24:13–18. doi: 10.1093/molbev/msl149. [DOI] [PubMed] [Google Scholar]
- 35.Hazkani-Covo E. Mitochondrial insertions into primate nuclear genomes suggest the use of numts as a tool for phylogeny. Mol Biol Evol. 2009;26:2175–2179. doi: 10.1093/molbev/msp131. [DOI] [PubMed] [Google Scholar]
- 36.Hazkani-Covo E., Zeller R.M., Martin W. Molecular poltergeists: mitochondrial DNA copies (numts) in sequenced nuclear genomes. PLoS Genet. 2010;6:e1000834. doi: 10.1371/journal.pgen.1000834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hodgkinson A., Idaghdour Y., Gbeha E., Grenier J.C., Hip-Ki E., Bruat V. High resolution genomic analysis of human mitochondrial RNA sequence variation. Science. 2014;344:413–415. doi: 10.1126/science.1251110. [DOI] [PubMed] [Google Scholar]
- 38.Hu Q.X., Fan Y., Xu L., Pang W., Wang S., Zheng Y.T. Analysis of the complete mitochondrial genome and characterization of diverse NUMTs of Macaca leonina. Gene. 2015;571:279–285. doi: 10.1016/j.gene.2015.06.085. [DOI] [PubMed] [Google Scholar]
- 39.Igloi G.L., Aldinger C.A. Where have all the inosines gone? Conflicting evidence for A-to-I editing of the anticodon of higher eukaryotic tRNAACGArg questions the dogma of a universal wobble-mediated decoding of CGN codons. IUBMB Life. 2016;68:419–422. doi: 10.1002/iub.1497. [DOI] [PubMed] [Google Scholar]
- 40.Jensen-Seaman M.I., Wildschutte J.H., Soto-Calderón I.D., Anthony N.M. A comparative approach shows differences in patterns of numt insertion during hominoid evolution. J Mol Evol. 2009;68:688–699. doi: 10.1007/s00239-009-9243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Käll L., Storey J.D., MacCoss M.J., Noble W.S. Posterior error probabilities and false discovery rates: two sides of the same coin. J Proteome Res. 2008;7:40–44. doi: 10.1021/pr700739d. [DOI] [PubMed] [Google Scholar]
- 42.Kim S.-H., Park Y.-Y., Yoo Y.-S., Cho H. Self-clearance mechanism of mitochondrial E3 ligase MARCH5 contributes to mitochondria quality control. FEBS J. 2015 doi: 10.1111/febs.13568. [DOI] [PubMed] [Google Scholar]
- 43.Klobutcher L.A., Farabaugh P.J. Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell. 2002;111:763–766. doi: 10.1016/s0092-8674(02)01138-8. [DOI] [PubMed] [Google Scholar]
- 44.Koley S., Adhya S. A voltage-gated pore for translocation of tRNA. Biochem Biophys Res Commun. 2013;439:23–29. doi: 10.1016/j.bbrc.2013.08.036. [DOI] [PubMed] [Google Scholar]
- 45.Korpelainen H. The evolutionary processes of mitochondrial and cholorplast genomes differ from those of nuclear genomes. Naturwissenschaften. 2004;91:505–511. doi: 10.1007/s00114-004-0571-3. [DOI] [PubMed] [Google Scholar]
- 46.Lee H.R., Johnson K.A. Fidelity of the human mitochondrial DNA polymerase. J Biol Chem. 2006;281:36236–36240. doi: 10.1074/jbc.M607964200. [DOI] [PubMed] [Google Scholar]
- 47.Li M., Wang I.X., Li Y., Bruzel A., Richards A.L., Toung J.M. Widespread RNA and DNA sequence differences in the human transcriptome. Science. 2011;333:53–58. doi: 10.1126/science.1207018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lopez J.V., Cevario S., O'Brien S.J. Complete nucleotide sequences of the domestic cat (Felis catus) mitochondrial genome and a transposed mtDNA tandem repeat (Numt) in the nuclear genome. Genomics. 1996;33:229–246. doi: 10.1006/geno.1996.0188. [DOI] [PubMed] [Google Scholar]
- 49.Lopez J.V., Cumver M., Stephens J.C., Johnoson W.E., O'Brien S.J. Rates of nuclear and cytoplasmic mitochondrial DNA sequence divergence in mammals. Mol Biol Evol. 1997;14:277–286. doi: 10.1093/oxfordjournals.molbev.a025763. [DOI] [PubMed] [Google Scholar]
- 50.Lopez J.V., Stephens J.C., O'Brien S.J. The long and short nuclear mitochondrial DNA (Numt) lineages. Trends Ecol Evol. 1997;12:114. doi: 10.1016/s0169-5347(97)84925-7. [DOI] [PubMed] [Google Scholar]
- 51.Lopez J.V., Yukhi N., Masuda R., Modi W., O'Brien S.J. Numt, a recent transfer and tandem amplification of mitochondrial DNA to the nuclear genome of the domestic cat. J Mol Evol. 1994;39:174–190. doi: 10.1007/BF00163806. [DOI] [PubMed] [Google Scholar]
- 52.Mao X., Dong J., Hua P., He G., Zhang S., Rossiter S.J. Heteroplasmy and ancient translocation of mitochondrial DNA to the nucleus in the Chinese Horseshoe Bat (Rhinolophus sinicus) complex. PLoS One. 2014;9:e98035. doi: 10.1371/journal.pone.0098035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Martinez-Rodriguez L., Erdogan O., Jimenez-Rodriguez M., Gonzalez-Rivera K., Williams T., Li L. Functional class I and II amino acid-activating enzymes can be coded by opposite strands of the same gene. J Biol Chem. 2015;290:19710–19725. doi: 10.1074/jbc.M115.642876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Michalovova M., Vyskot B., Kejnovsky E. Analysis of plastid and mitochondrial DNA insertions in the nucleus (NUPTs and NUMTs) of six plant species: size, relative age and chromosomal localization. Heredity. 2013;111:314–320. doi: 10.1038/hdy.2013.51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Michel C.J. Circular code motifs in transfer and 16S ribosomal RNAs: a possible translation code in genes. Comput Biol Chem. 2012;45:17–29. doi: 10.1016/j.compbiolchem.2011.10.002. [DOI] [PubMed] [Google Scholar]
- 56.Michel C.J. An extended genetic scale of reading frame coding. J Theor Biol. 2015;365:164–174. doi: 10.1016/j.jtbi.2014.09.040. [DOI] [PubMed] [Google Scholar]
- 57.Michel C.J. The maximal C(3) self-complementary trinucleotide circular code X in genes of bacteria, eukaryotes, plasmids and viruses. J Theor Biol. 2015;380:156–177. doi: 10.1016/j.jtbi.2015.04.009. [DOI] [PubMed] [Google Scholar]
- 58.Michel C.J., Seligmann H. Bijective transformation circular codes and nucleotide exchanging RNA transcription. Biosystems. 2014;118:39–50. doi: 10.1016/j.biosystems.2014.02.002. [DOI] [PubMed] [Google Scholar]
- 59.Moreira S., Valach M., Aoulad-Aissa M., Otto C., Burger G. Novel modes of RNA editing in mitochondria. Nucleic Acids Res. 2016;44:4907–4919. doi: 10.1093/nar/gkw188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Frenkel-Morgenstern M., Gorohovski A., Lacroix V., Rogers M., Ibanez K., Boullosa C. ChiTaRS: a database of human, mouse and fruit fly chimeric transcripts and RNA-sequencing data. Nucleic Acids Res. 2013;41:D142–D151. doi: 10.1093/nar/gks1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Ojala D., Montoya J., Attardi G. tRNA punctuation model of RNA processiong in human mitochondria. Nature. 1981;290:470–474. doi: 10.1038/290470a0. [DOI] [PubMed] [Google Scholar]
- 62.Olson L.E., Yoder A.D. Using secondary structure to identify ribosomal numts: cautionary examples from the human genome. Mol Biol Evol. 2002;19:93–100. doi: 10.1093/oxfordjournals.molbev.a003987. [DOI] [PubMed] [Google Scholar]
- 63.Ramos A., Barbena E., Mateiu L., del Mar González M., Mairal Q., Lima M. Nuclear insertions of mitochondrial origin: database updating and usefulness in cancer studies. Mitochondrion. 2011;11:946–953. doi: 10.1016/j.mito.2011.08.009. [DOI] [PubMed] [Google Scholar]
- 64.Ramos A., Santos C., Mateiu L., del Mar Gonzalez M., Alvarez L., Azevedo L. Frequency and pattern of heteroplasmy in the complete human mitochondrial genome. PLoS One. 2013;8:e74636. doi: 10.1371/journal.pone.0074636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ren T., Liang S., Zhao A., He K. Analysis of the complete mitochondrial genome of the Zhedong White goose and characterization of NUMTs: reveal domestication history of goose in China and Euro. Gene. 2016;577:75–81. doi: 10.1016/j.gene.2015.11.018. [DOI] [PubMed] [Google Scholar]
- 66.Ricchetti M., Tekala F., Dujon B. Continued colonization of the human genome by mitochondrial DNA. PLoS Biol. 2004;2:E273. doi: 10.1371/journal.pbio.0020273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Richly E., Leister D. NUMTs in sequenced eukaryotic genomes. Mol Biol Evol. 2004;21:1081–1084. doi: 10.1093/molbev/msh110. [DOI] [PubMed] [Google Scholar]
- 68.Riddle D.L., Carbon J. Frameshift suppression: a nucleotide addition in the anticodon of a glycine transfer RNA. Nat New Biol. 1973;242:230–234. doi: 10.1038/newbio242230a0. [DOI] [PubMed] [Google Scholar]
- 69.Rodin S.N., Ohno S. 2 Types of aminoacyl-transfer-RNA synthetases could be originally encoded by complementary strands of the same nucleic-acid. Orig Life Evol Biosph. 1995;23:393–418. doi: 10.1007/BF01582025. [DOI] [PubMed] [Google Scholar]
- 70.Rodin S., Rodin A., Ohno S. The presence of codon–anticodon pairs in the acceptor stem of tRNAs. Proc Natl Acad Sci U S A. 1996;93:4537–4542. doi: 10.1073/pnas.93.10.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rogers H.H., Griffiths-Jones S. Mitochondrial pseudogenes in the nuclear genomes of Drosophila. PLoS One. 2012;7:e32593. doi: 10.1371/journal.pone.0032593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Root-Bernstein M., Root-Bernstein R. The ribosome as a missing link in the evolution of life. J Theor Biol. 2015;367:130–158. doi: 10.1016/j.jtbi.2014.11.025. [DOI] [PubMed] [Google Scholar]
- 73.Root-Bernstein M., Root-Bernstein R. The ribosome as a missing link in prebiotic evolution II: ribosomes encode ribosomal proteins that bind to common regions of their own mRNAs and rRNAs. J Theor Biol. 2016;397:115–127. doi: 10.1016/j.jtbi.2016.02.030. [DOI] [PubMed] [Google Scholar]
- 74.Rose G., Passarino G., Scornaienchi V., Romeo G., Dato S., Bellizzi D. The mitochondrial DNA control region shows genetically correlated levels of heteroplasmy in leukocytes of centenarians and their offspring. BMC Genomics. 2007;8:293. doi: 10.1186/1471-2164-8-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Rubio M.A., Hopper A.K. Transfer RNA travels from the cytoplasm to organelles. Wiley Interdiscip Rev RNA. 2011;2:802–817. doi: 10.1002/wrna.93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Salinas T., Duby F., Larosa V., Coosemans N., Bonnefoy N., Motte P. Co-evolution of mitochondrial tRNA import and codon usage determines translational efficiency in the green alga Chlamydomonas. PLoS Genet. 2012;8:e1002946. doi: 10.1371/journal.pgen.1002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Schmitz J., Piskurek O., Zischler H. Forty million years of independent evolution: a mitochondrial gene and its corresponding nuclear pseudogene in primates. J Mol Evol. 2005;61:1–11. doi: 10.1007/s00239-004-0293-3. [DOI] [PubMed] [Google Scholar]
- 78.Schneider A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu Rev Biochem. 2011;80:1033–1053. doi: 10.1146/annurev-biochem-060109-092838. [DOI] [PubMed] [Google Scholar]
- 79.Seligmann H. Hybridization between mitochondrial heavy strand tDNA and expressed light strand tRNA modulates the function of heavy strand tDNA as light strand replication origin. J Mol Biol. 2008;379:188–199. doi: 10.1016/j.jmb.2008.03.066. [DOI] [PubMed] [Google Scholar]
- 80.Seligmann H. Mitochondrial tRNAs as light strand replication origins: similarity between anticodon loops and the loop of the light strand replication origin predicts initiation of DNA replication. Biosystems. 2010;99:85–93. doi: 10.1016/j.biosystems.2009.09.003. [DOI] [PubMed] [Google Scholar]
- 81.Seligmann H. Avoidance of antisense, antiterminator tRNA anticodons in vertebrate mitochondria. Biosystems. 2010;101:42–50. doi: 10.1016/j.biosystems.2010.04.004. [DOI] [PubMed] [Google Scholar]
- 82.Seligmann H. Undetected antisense tRNAs in mitochondrial genomes? Biol Direct. 2010;5:39. doi: 10.1186/1745-6150-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Seligmann H. Pathogenic mutations in antisense mitochondrial tRNAs. J Theor Biol. 2011;269:287–296. doi: 10.1016/j.jtbi.2010.11.007. [DOI] [PubMed] [Google Scholar]
- 84.Seligmann H. Two genetic codes, one genome: frameshifted primate mitochondrial genes code for additional proteins in presence of antisense tRNAs. Biosystems. 2011;105:271–285. doi: 10.1016/j.biosystems.2011.05.010. [DOI] [PubMed] [Google Scholar]
- 85.Seligmann H. An overlapping genetic code for frameshifted overlapping genes in Drosophila mitochondria: antisense antitermination tRNAs UAR insert serine. J Theor Biol. 2012;298:51–76. doi: 10.1016/j.jtbi.2011.12.026. [DOI] [PubMed] [Google Scholar]
- 86.Seligmann H. Overlapping genetic codes for overlapping frameshifted genes in Testudines, and Lepidochelys olivacea as special case. Comput Biol Chem. 2012;41:18–34. doi: 10.1016/j.compbiolchem.2012.08.002. [DOI] [PubMed] [Google Scholar]
- 87.Seligmann H. Coding constraints modulate chemically spontaneous mutational replication gradients in mitochondrial genomes. Curr Genomics. 2012;13:37–54. doi: 10.2174/138920212799034802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Seligmann H. Overlapping genes coded in the 3′-to-5′-direction in mitochondrial genes and 3′-to-5′ polymerization of non-complementary RNA by an 'invertase'. J Theor Biol. 2012;315:38–52. doi: 10.1016/j.jtbi.2012.08.044. [DOI] [PubMed] [Google Scholar]
- 89.Seligmann H. Putative mitochondrial polypeptides coded by expanded quadruplet codons decoded by antisense tRNAs with unusual anticodons. Biosystems. 2012;110:84–106. doi: 10.1016/j.biosystems.2012.09.002. [DOI] [PubMed] [Google Scholar]
- 90.Seligmann H. Pocketknife tRNA hypothesis: anticodons in mammal mitochondrial tRNA side-arm loops translate proteins? Biosystems. 2013;113:165–176. doi: 10.1016/j.biosystems.2013.07.004. [DOI] [PubMed] [Google Scholar]
- 91.Seligmann H. Putative protein-encoding genes within mitochondrial rDNA and the D-Loop region. In: Lin Z., Liu W., editors. Ribosomes: molecular structure, role in biological functions and implications for genetic diseases. 2013. pp. 67–86. [chapter 4] [Google Scholar]
- 92.Seligmann H. Triplex DNA:RNA, 3′-to-5′ inverted RNA and protein coding in mitochondrial genomes. J Comput Biol. 2013;20:660–671. doi: 10.1089/cmb.2012.0134. [DOI] [PubMed] [Google Scholar]
- 93.Seligmann H. Polymerization of non-complementary RNA: systematic symmetric nucleotide exchanges mainly involving uracil produce mitochondrial transcripts coding for cryptic overlapping genes. Biosystems. 2013;111:156–174. doi: 10.1016/j.biosystems.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 94.Seligmann H. Systematic asymmetric nucleotide exchanges produce human mitochondrial RNAs cryptically encoding for overlapping protein coding genes. J Theor Biol. 2013;324:1–20. doi: 10.1016/j.jtbi.2013.01.024. [DOI] [PubMed] [Google Scholar]
- 95.Seligmann H. Putative anticodons in mitochondrial sidearm loops: Pocketknife tRNAs? J Theor Biol. 2014;340:155–163. doi: 10.1016/j.jtbi.2013.08.030. [DOI] [PubMed] [Google Scholar]
- 96.Seligmann H. Mitochondrial swinger replication: DNA replication systematically exchanging nucleotides and short 16S ribosomal DNA swinger inserts. Biosystems. 2014;25:22–31. doi: 10.1016/j.biosystems.2014.09.012. [DOI] [PubMed] [Google Scholar]
- 97.Seligmann H. Species radiation by DNA replication that systematically exchanges nucleotides? J Theor Biol. 2014;363:216–222. doi: 10.1016/j.jtbi.2014.08.036. [DOI] [PubMed] [Google Scholar]
- 98.Seligmann H. Phylogeny of genetic codes and punctuation codes within genetic codes. Biosystems. 2015;129:36–43. doi: 10.1016/j.biosystems.2015.01.003. [DOI] [PubMed] [Google Scholar]
- 99.Seligmann H. Sharp switches between regular and swinger mitochondrial replication: 16S rDNA systematically exchanging nucleotides A ↔ T + C ↔ G in the mitogenome of Kamimuria wangi. Mitochondrial DNA. 2016;27:2440–2446. doi: 10.3109/19401736.2015.1033691. [DOI] [PubMed] [Google Scholar]
- 100.Seligmann H. Swinger RNAs with sharp switches between regular transcription and transcription systematically exchanging ribonucleotides: case studies. Biosystems. 2015;135:1–8. doi: 10.1016/j.biosystems.2015.07.003. [DOI] [PubMed] [Google Scholar]
- 101.Seligmann H. Systematic exchanges between nucleotides: genomic swinger repeats and swinger transcription in human mitochondria. J Theor Biol. 2015;384:70–77. doi: 10.1016/j.jtbi.2015.07.036. [DOI] [PubMed] [Google Scholar]
- 102.Seligmann H. Codon expansion and systematic transcriptional deletions produce tetra-, pentacoded mitochondrial peptides. J Theor Biol. 2015;387:154–165. doi: 10.1016/j.jtbi.2015.09.030. [DOI] [PubMed] [Google Scholar]
- 103.Seligmann H. Translation of mitochondrial swinger RNAs according to tri-, tetra- and pentacodons. Biosystems. 2016;140:38–48. doi: 10.1016/j.biosystems.2015.11.009. [DOI] [PubMed] [Google Scholar]
- 104.Seligmann H. Swinger RNA self-hybridization and mitochondrial non-canonical swinger transcription, transcription systematically exchanging nucleotides. J Theor Biol. 2016;399:84–91. doi: 10.1016/j.jtbi.2016.04.007. [DOI] [PubMed] [Google Scholar]
- 105.Seligmann H. Systematically frameshifting by deletion of every 4th or 4th and 5th nucleotides during mitochondrial transcription: RNA self-hybridization regulates delRNA expression. Biosystems. 2016;142:43–51. doi: 10.1016/j.biosystems.2016.03.009. [DOI] [PubMed] [Google Scholar]
- 106.Seligmann H., Krishnan N.M. Mitochondrial replication origin stability and propensity of adjacent tRNA genes to form putative replication origins increase developmental stability in lizards. J Exp Zool B Mol Dev Evol. 2006;306:433–449. doi: 10.1002/jez.b.21095. [DOI] [PubMed] [Google Scholar]
- 107.Seligmann H., Krishnan N.M., Rao B.J. Possible multiple origins of replication in primate mitochondria: alternative role of tRNA sequences. J Theor Biol. 2006;241:321–332. doi: 10.1016/j.jtbi.2005.11.035. [DOI] [PubMed] [Google Scholar]
- 108.Seligmann H., Krishnan N.M., Rao B.J. Mitochondrial tRNA sequences as unusual replication origins: pathogenic implications for Homo sapiens. J Theor Biol. 2006;243:375–385. doi: 10.1016/j.jtbi.2006.06.028. [DOI] [PubMed] [Google Scholar]
- 109.Seligmann H., Labra A. Tetracoding increases with body temperature in Lepidosauria. Biosystems. 2013;447:155–163. doi: 10.1016/j.biosystems.2013.09.002. [DOI] [PubMed] [Google Scholar]
- 110.Seligmann H., Labra A. The relation between hairpin formation by mitochondrial WANCY tRNAs and the occurrence of the light strand replication origin in Lepidosauria. Gene. 2014;542:248–257. doi: 10.1016/j.gene.2014.02.021. [DOI] [PubMed] [Google Scholar]
- 111.Seligmann H., Raoult D. Unifying view of stem-loop hairpin RNA as origin of current and ancient parasitic and non-parasitic RNAs, including in giant viruses. Curr Opin Microbiol. 2016;31:1–8. doi: 10.1016/j.mib.2015.11.004. [DOI] [PubMed] [Google Scholar]
- 112.Sharma A., Sharma A. Plasmodium falciparum mitochondria import tRNAs along with an active phenylalanyl-tRNA synthetase. Biochem J. 2015;465:459–469. doi: 10.1042/BJ20140998. [DOI] [PubMed] [Google Scholar]
- 113.Shi H., Dong J., Irwin D.M., Zhang S., Mao X. Repetitive transpositions of mitochondrial DNA sequences to the nucleus during the radiation of horseshoe bats (Rhinolophus, Chiroptera) Gene. 2016;581:161–169. doi: 10.1016/j.gene.2016.01.035. [DOI] [PubMed] [Google Scholar]
- 114.Sieber F., Duchêne A.M., Maréchal-Drouard L. Mitochondrial RNA import: from diversity of natural mechanisms to potential applications. Int Rev Cell Mol Biol. 2011;287:145–190. doi: 10.1016/B978-0-12-386043-9.00004-9. [DOI] [PubMed] [Google Scholar]
- 115.Smigrodzki R.M., Khan S.M. Mitochondrial microheteroplasmy and a theory of aging and age-related disease. Rejuvenation Res. 2005;8:172–198. doi: 10.1089/rej.2005.8.172. [DOI] [PubMed] [Google Scholar]
- 116.Song S., Jiang F., Yuan J., Guo W., Miao Y. Exceptionally high cumulative percentage of NUMTs originating from linear mitochondrial DNA molecules in the Hydra magnipapillata genome. BMC Genomics. 2013;14:447. doi: 10.1186/1471-2164-14-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Song H., Moulton M.J., Whiting M.F. Rampant nuclear insertion of mtDNA across diverse lineages within Orthoptera (Insecta) PLoS One. 2014;9:e110508. doi: 10.1371/journal.pone.0110508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Soto-Calderón I.D., Clark N.J., Wildschutte J.V., DiMattio K., Jensen-Seaman M.I., Anthony N.M. Identification of species-specific nuclear insertions of mitochondrial DNA (numts) in gorillas and their potential as population genetic markers. Mol Phylogenet Evol. 2014;81:61–70. doi: 10.1016/j.ympev.2014.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Soto-Calderón I.D., Lee E.J., Jensen-Seaman M.I., Anthony N.M. Factors affecting the relative abundance of nuclear copies of mitochondrial DNA (numts) in hominoids. J Mol Evol. 2012;75:102–111. doi: 10.1007/s00239-012-9519-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Sroga G.E., Nemoto F., Kuchino Y., Bjork G.R. Insertion (sufB) in the anticodon loop or base substitution (sufC) in the anticodon stem of tRNA(Pro)2 from Salmonella typhimurium induces suppression of frameshift mutations. Nucleic Acids Res. 1992;20:3463–3469. doi: 10.1093/nar/20.13.3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Stefano G.B., Kream R.M. Mitochondrial DNA heteroplasmy in human health and disease. Biomed Rep. 2016;4:259–262. doi: 10.3892/br.2016.590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Taylor E.B., Rutter J. Mitochondrial quality control by the ubiquitin-prooteasome system. Biochem Soc Trans. 2011;39:1509–1513. doi: 10.1042/BST0391509. [DOI] [PubMed] [Google Scholar]
- 123.Thalman O., Hebler J., Poinar H.N., Päabo S., Vigilant L. Unreliable mtDNA data due to nuclear insertions: a cautionary tale from analysis of humans and other great apes. Mol Ecol. 2004;13:321–335. doi: 10.1046/j.1365-294x.2003.02070.x. [DOI] [PubMed] [Google Scholar]
- 124.Thalman O., Serre D., Hofreiter M., Lukas D., Eriksson J., Vigilant L. Nuclear insertions help and hinder inference of the evolutionary history of gorilla mtDNA. Mol Ecol. 2005;14:179–188. doi: 10.1111/j.1365-294X.2004.02382.x. [DOI] [PubMed] [Google Scholar]
- 125.Tourmen Y., Baris O., Dessen P., Jacques C., Malthièry Y., Reynier P. Structure and chromosomal distribution of human mitochondrial pseudogenes. Genomics. 2002;80:71–77. doi: 10.1006/geno.2002.6798. [DOI] [PubMed] [Google Scholar]
- 126.Tsuji J., Frith M.C., Tomii K., Horton P. Mammalian NUMT insertion is non-random. Nucleic Acids Res. 2012;40:9073–9088. doi: 10.1093/nar/gks424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Tuohy T.M., Thompson S., Gesteland R.F., Atkins J.F. Seven, eight and nine-membered anticodon loop mutants of tRNA(2Arg) which cause + 1 frameshifting. Tolerance of DHU arm and other secondary mutations. J Mol Biol. 1992;228:1042–1054. doi: 10.1016/0022-2836(92)90313-9. [DOI] [PubMed] [Google Scholar]
- 128.Verscheure S., Backeljau T., Desmyter S. In silico discovery of a nearly complete mitochondrial genome Numt in the dog (Canis lupus familiaris) nuclear genome. Genetica. 2015;143:453–458. doi: 10.1007/s10709-015-9844-3. [DOI] [PubMed] [Google Scholar]
- 129.Wang B., Zhou X., Shi F., Liu Z., Roos C., Garber P.A. Full-length Numt analysis provides evidence for hybridization between the Asian colobine genera Trachypithecus and Semnopithecus. Am J Primatol. 2015;77:901–910. doi: 10.1002/ajp.22419. [DOI] [PubMed] [Google Scholar]
- 130.Wang I.X., Core L.J., Kwak H., Brady L., Bruzel A., McDaniel L. RNA-DNA differences are generated in human cells within seconds after RNA exits polymerase II. Cell Rep. 2014;6:906–915. doi: 10.1016/j.celrep.2014.01.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Xie B., Yang W., Chen L., Jiang H., Liao Y., Liao D.J. Two RNAs or DNAs may artificially fuse together at a short homologous sequence (SHS) during reverse transcription or polymerase chain reactions, and thus reporting an SHS-containing chimeric RNA requires extra caution. PLoS One. 2016;11:e0154855. doi: 10.1371/journal.pone.0154855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Yang W., Wu J.-M., Bi A.-D., Ou-yang Y., Shen H.-H., Chim G.-W. Possible formation of mitochondrial-RNA containing chimeric or trimeric RNA implies a post-transcriptional and post-splicing mechanism for RNA fusion. PLoS One. 2013;8:e77016. doi: 10.1371/journal.pone.0077016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Yao Y.G., Kong Q.P., Salas A., Bandelt H.J. Pseudomitochondrial genome haunts disease studies. J Med Genet. 2008;45:769–772. doi: 10.1136/jmg.2008.059782. [DOI] [PubMed] [Google Scholar]
- 134.Zhang D.X., Hewitt G.M. The long and short of nuclear mitochondrial DNA (Numt) lineages Reply from D-X. Zhang and G.M. Hewitt. Trends Ecol Evol. 1997;12:114. doi: 10.1016/s0169-5347(97)84926-9. [DOI] [PubMed] [Google Scholar]