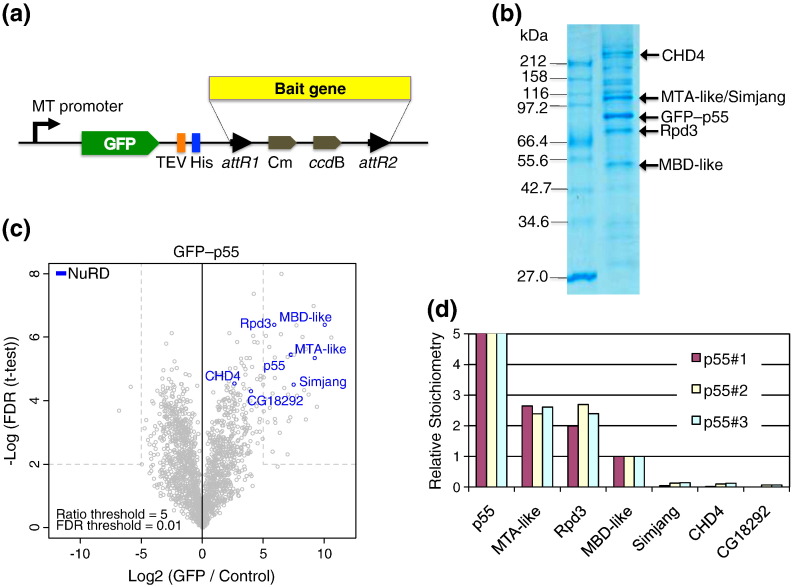
Fig. 1.
Endogenous dNuRD complex from Drosophila. (a) The endogenous dNuRD complex was purified from Drosophila S2 Schneider cells using affinity capture of a GFP-tagged bait protein. The construct used for expressing tandem affinity purification (TAP) tagged bait protein is shown in a schematic fashion. MT, matellothionin promoter; GFP, green fluorescent protein; TEV, tobacco etch virus NIa protease recognition sequence; His, oligohistidine tag; attR1/attR2, recombination sites; Cm, chloramphenicol resistance marker; ccdB, selection cassette. (b) The proteins captured by GFP-trap resin (Chromotek) from a nuclear extract of cells expressing GFP-p55 were analyzed by SDS-PAGE and Coommassie brilliant blue (CBB) staining. Components identified by MS and Western blot are marked with arrows and denoted. (c) Statistically enriched proteins in the GFP-p55 pull-down experiment were identified in a permutation-based FDR-corrected t test and are shown in a Volcano plot. (d) The relative stoichiometries of components within endogenous dNuRD were determined using intensity-based absolute quantification [23]. The values are normalized to MBD-like (stoichiometry = 1.0). Three individually purified samples (denoted p55#1, p55#2, and p55#3) consistently indicate a ratio of 2: 2: 1 for the NuRD core components MTA-like, Rpd3, and MBD-like. In contrast, CHD4, Simjang, and CG18292 appear to bind sub-stoichiometrically.