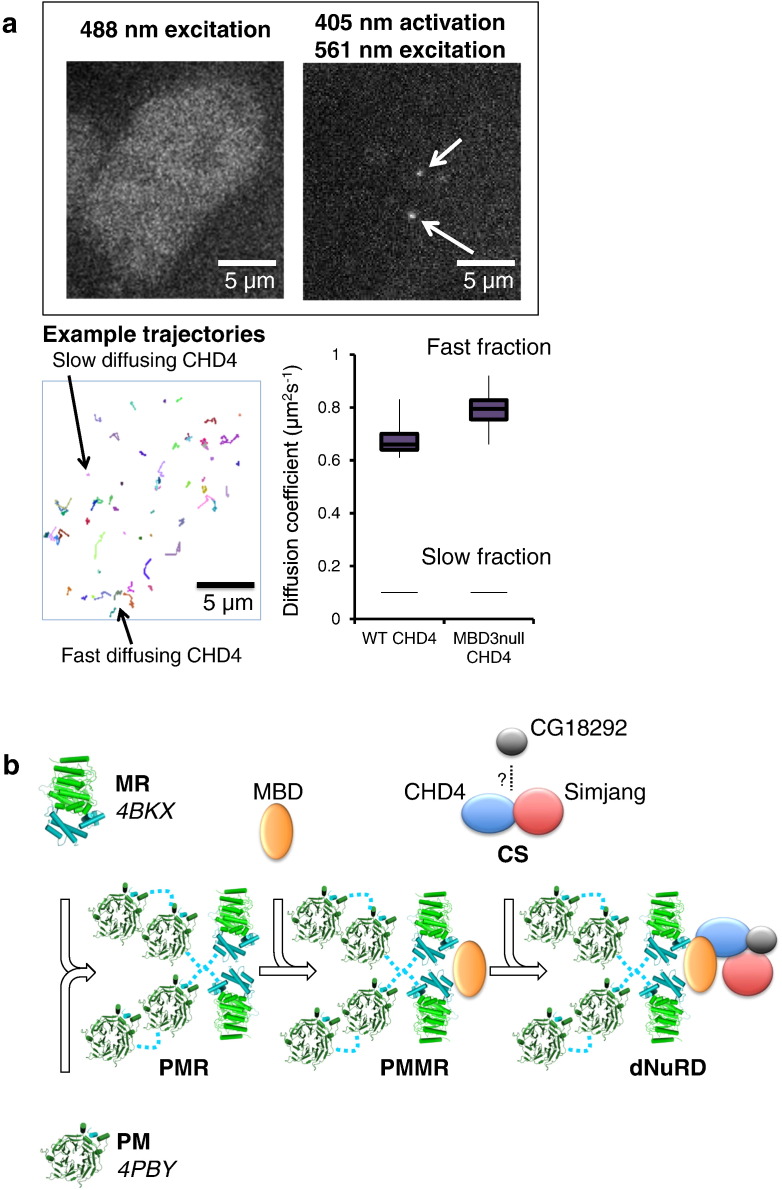
Fig. 5.
Interaction of CHD4 with the core PMMR complex. (a) 2D single-molecule tracking of single mEos3-tagged CHD4 molecules in live mESCs show differences in the diffusion of a sub-population of CHD4 molecules in the presence (WT) and absence of MBD3 (MBD3-null). Representative images of the same cell are shown (top right) using low power 488-nm excitation (green form of mEos3) and 405-nm/561-nm excitation (photo-activated red form of mEos3). A small number of the individual tracks from this cell are also shown indicating the fast and slow diffusing fractions of CHD4. The exact diffusion coefficients extracted from the data are shown in a box-and-whisker plot (lower right). A total of 23,854 and 20,039 tracks were analyzed for the wild-type and MBD3-null cells, respectively. (b) A putative model of NuRD complex assembly is shown in a schematic representation. Protein interactions which can be modeled from known molecular structures (PDB identifiers 4BKX and 4PBY) are shown. The interactions between MTA-like (light blue) and Rpd3 (light green) (MR) are based on the structure of MTA1/HDAC1 elucidated by X-ray crystallography (PDB ID 4BKX, [15]). The interaction of p55 (dark green) with MTA-like is modeled on the structure of RbAp48 with a C-terminal fragment of MTA1 (PDB ID 4PBY, [16]). Two molecules of p55 interact with one molecule of MTA-like via interactions with two related peptide motifs in the centre and C-terminus of MTA-like (residues 618–622 KKAARQ and 848–853 RRAARK, respectively). The p55, MTA-like, and Rpd3 proteins assemble into a complex with presumed 4:2:2 stoichiometry (PMR). One copy of MBD-like protein (yellow) interacts with PMR to yield the core NuRD complex (PMMR) with a stoichiometry of 4:2:1:2 consistent with that determined by SEC-MALLS (Supplementary Fig. S2). Simjang (dark blue), CHD4 (red), and CG18292 (putative Doc1, gray) are more loosely associated, peripheral subunits that interact with the core NuRD complex. CHD4 does not interact with core PMMR by itself, but likely interacts as a sub-module with Simjang (and perhaps CG18292) via the interaction of Simjang with MBD-like (as structurally characterized by the p66α–MBD2 interaction), to give holo-NuRD. The interaction of the two sub-modules combines the ATP-dependent chromatin remodeling function of CHD4 with the deacetylase activity conferred by Rpd3 in one holo-enzyme. Preformed PMR and PMMR sub-modules (and quite possibly MR) represent stable and enzymatically active deacetylases, which may catalyze distinct cellular functions on their own.