Figure 6.
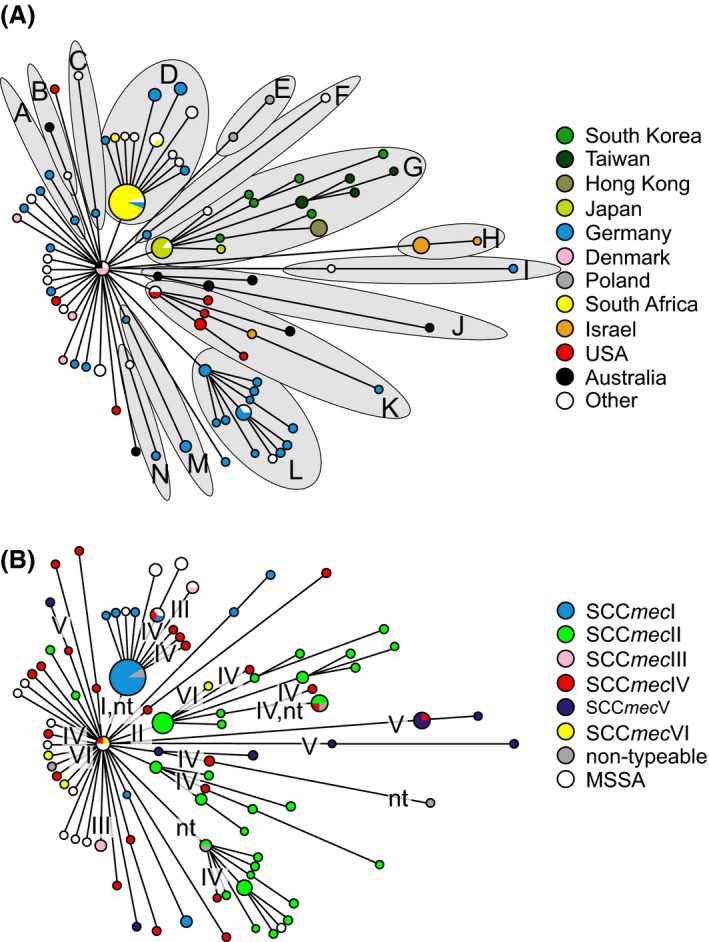
Minimum‐spanning tree constructed from core genomic sequences of 135 isolates of the ST5 Staphylococcus aureus clone collected across the globe. Figure reproduced from fig. 1A,B of Nübel et al. (2008), Copyright (2008) National Academy of Sciences, USA. The sizes of circles are proportional to the haplotype frequency, and the line length is proportional to the number of mutational steps between haplotypes. In panel (A) haplotypes are painted with colours in proportion to the country of origin that isolates with this haplotype were found. Fourteen lineages (labelled ‘A' through ‘N') tend to cluster with geographical origin emerge, suggesting that specific haplotypes of the core genome of the S. aureus ST5 clone are largely endemic to a geographical area. In panel (B) the same minimum‐spanning tree is used, but now haplotypes are painted with colours in proportion to the SCCmec element type co‐occurring in the isolate. Some SCCmec elements are nontypeable (‘nt'), likely due to being novel variants. The most parsimonious distribution of acquisition events of SCCmec is indicated with roman numerals placed along the branches of the tree (where the roman numeral corresponds to the SCCmec type), although the actual number of SCCmec acquisitions may be higher. Note that while the different SCCmec types are present across the globe, the acquisition events tend to occur at the tips of the tree, suggesting that methicillin resistance tends to evolve within a local S. aureus strain as opposed to occurring once and then disseminating globally.
