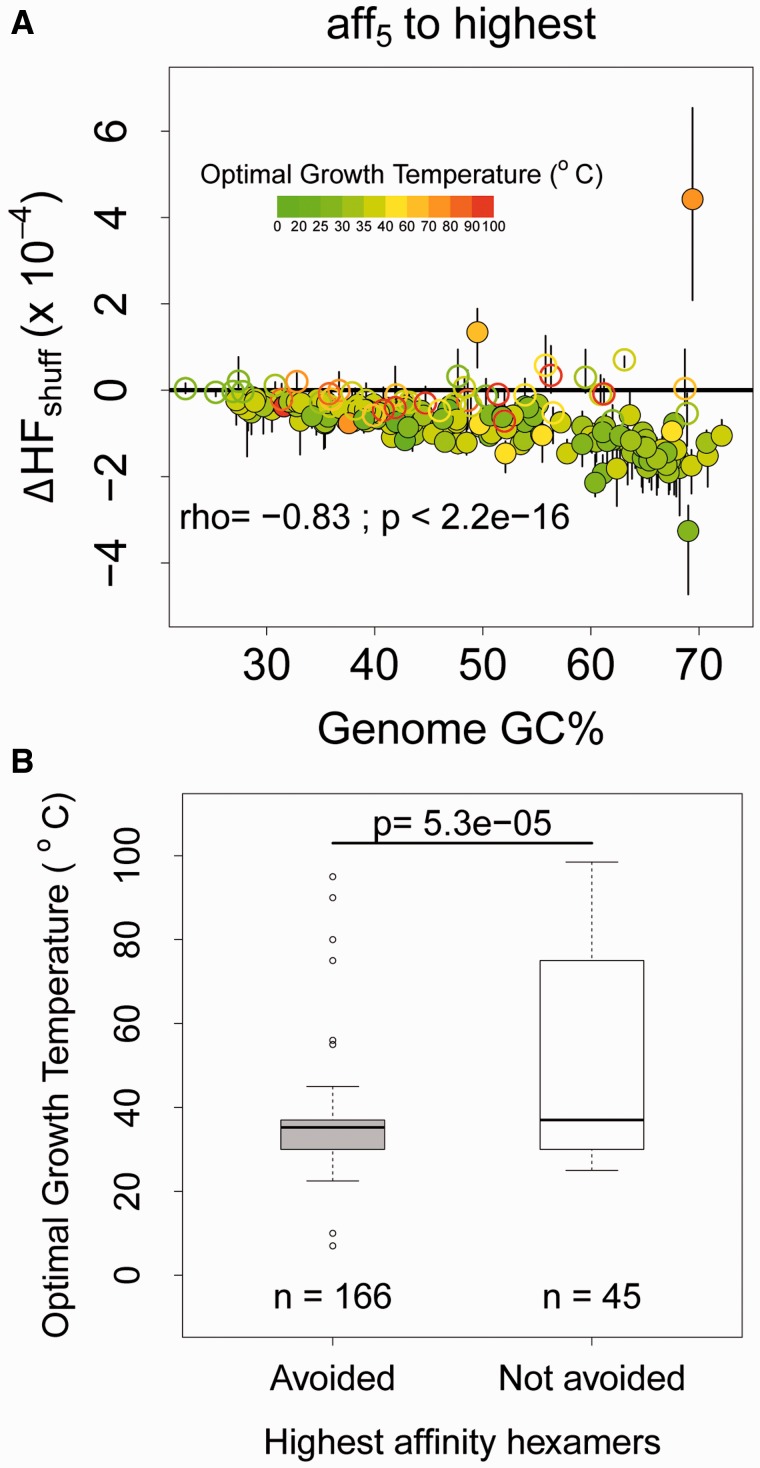
Fig. 3.—
(A) Median and interquartile range of corrected HF (ΔHFshuff = HFobs − HFexp) for hexamers as a function of genome GC%, for the highest affinity bin. The horizontal solid black line indicates no difference in observed and randomized HF. Points are colored according to the optimal growth temperature of the organism. Filled circles indicate that HFobs is significantly different from HFexp. Spearman’s rho and P-values are given for the relationship between ΔHF and genome GC% (for filled circles below 0). (B) Boxplots showing optimal growth temperatures for genomes that avoid SD-like hexamers versus genomes that do not (based on SD::anti-SD affinity calculations at the respective growth temperatures). The distributions are significantly different from each other (Wilcoxon rank sum test).